Covalent Heterocyclic Fragment Library for identification of Cryptic and Allosteric Pocket
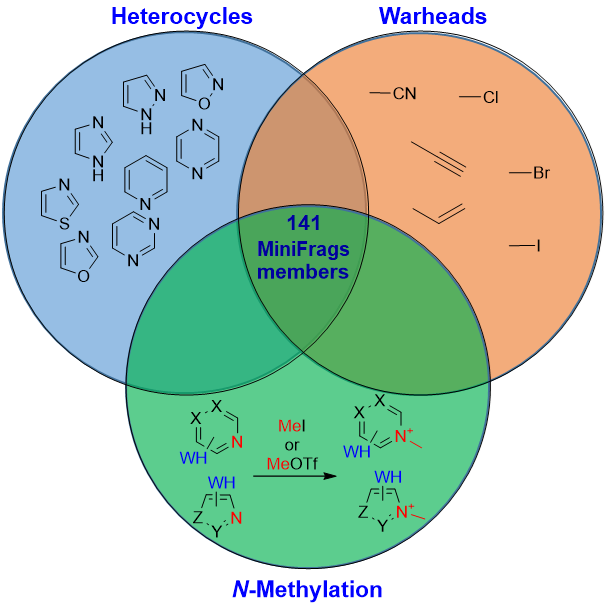
141 compounds
This unique library of small heterocyclic electrophiles developed by the research group of Prof. György Keserű have been shown to be effective in finding tiny new binding pockets for different protein targets (MedChemComm 2019, Nat. Commun. 2020 and Pharmaceuticals 2022). The library combines the advantages of Astex’s MiniFrags in exploring unprecedented binding site with that of covalent binders in higher affinity and easier detection. It consists of six and five-membered heterocycles most abundant in drugs that are equipped with only one or two-atom covalent warheads. This makes it unique in contrast to those often used in covalent screening, much larger acrylamides and chloroacetamides which can significantly influence the binding mode of active molecules. Thus, applying the smallest possible covalent function helps to avoid promiscuity and keep the same recognition pattern of non-covalent scaffolds.
On the other hand, the electrophile-first approach proved to be an effective way of developing covalent drugs (Nat. Rev. Drug Discov. 2022). A number of covalent probes and drug candidates, including recent examples of Nirmatrelvir and Sotorasib have been developed using covalent screening techniques.
Covalent MiniFrags is a unique tool for searching of new binding pockets, elaboration of discovered hits and growing vector identification. The XChem facility at Diamond LightSource UK is a strategic partner in pioneering applications of Covalent MiniFrags.
Typical Formats
Catalog No.
CMF-141-5-Y-500
Compounds
141
1 plate
Amount
5 μL of 500 mM DMSO solutions
Plates and formats
384-well acoustic LDV plates, first two and last two columns empty
Price
Catalog No.
CMF‑141‑25‑X‑100
Compounds
141
2 plates
Amount
25 μL of 100 mM DMSO solutions
Plates and formats
96-well plates, Greiner Cat. No 650201, round (U) bottom, 1 & 12 columns empty, 80 compounds per plate
Price
Download SD files
Key features
- Most common nitrogen-containing heterocycles
- The smallest covalent warheads
- Experimentally characterized stability and intrinsic reactivity
- Evaluated on a number of targets
Covalent MiniFrags Library consists of 85 heterocyclic electrophiles and 56 N-methylated functional heterocycles. Both subsets are described in detail in scientific publications and are now available for your research.
Specially selected molecules to target bromodomains
15 360 compounds
Bromodomains are protein domains found in various proteins and are involved in the recognition of acetylated lysine residues on histone proteins. These domains are named after their ability to recognize the acetyl-lysine side chain, which has a similar shape to a bromide ion. Bromodomains are essential in regulating gene expression and chromatin structure, as acetylation of histones is associated with open chromatin and active gene transcription. Dysregulation of bromodomain-containing proteins has been implicated in several diseases, including cancer and inflammation. As a result, bromodomains have become a popular target for drug discovery efforts, with several small molecule inhibitors in development for cancer and other diseases.
We focused on the search of active compounds against the most important Bromodomain families: BET subfamily: includes BRD2, BRD3, BRD4, and BRDT bromodomains, which are characterized by an extended ZA loop that interacts with acetylated lysine residues on histones. GCN5-related subfamily: This subfamily includes the GCN5 and PCAF bromodomains found in histone acetyltransferases and have a shorter ZA loop than the BET subfamily. TAF1-like subfamily: TAF1 and TAF1L bromodomains are found in transcription factor TFIID and have an N-terminal extension that interacts with other complex subunits. ATAD2-like subfamily: ATAD2 and ATAD2B bromodomains are found in AAA+ ATPases and have a long ZA loop and a unique insertion between the second and third alpha-helices. Additionally, we run docking calculations for BPTF protein that also contains bromodomain.
Typical Formats
Bromodomain Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
BRD-15-0-Z-10
Compounds
15 360
12 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
BRD-15-10-Y-10
Compounds
15 360
48 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
BRD-15-50-Y-10
Compounds
15 360
48 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
BRD-15-10-Y-10 screening library 15 360 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library code: BRD-15
Version: 7 December 2023
15 360 compounds
sublibrary of EPG-38080
Library design
We used a structure-based approach, molecular docking calculations, as the main method for the library design. All available PDB structures for each of the bromodomain subfamilies were analyzed and extracted from PDB and PDBe. Unique "protein-ligand" complexes were selected for analysis. The following PDB structures were included in our study: BRD2 (4j1p, 4a9o, 5uew, 6ffe, 7l6d, 7l9j, and 7oe8), BRD3 (3s91 and 7l9l), BRD4 (7ajn, 7axr, and 7c2z), and BRDT (4flp, 7l9a, and 7mrg); PCAF (5fdz and 5fe5); TAF1 (5i1q, 6p38, and 7jjg); ATAD2 (6veo); and BPTF (5r4i, 6lu5, and 7lp0).
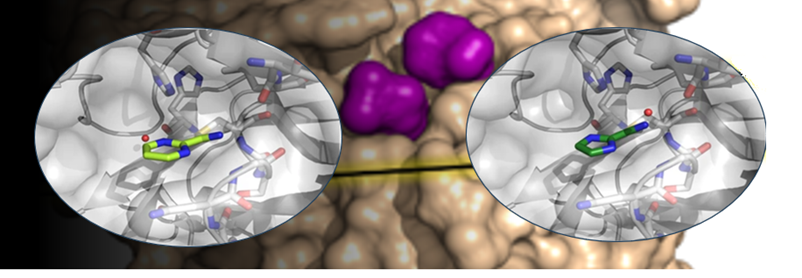
All simulations were performed with a common feature: the binding of a potential ligand to Asn inside the bromodomain binding pocket. All other binding points were dependent on the specific bromodomain and the structure of its native ligand. For example, in the left picture, the docking example of 7l6d is presented. The model is characterized by the following key features: (1) an h-bond acceptor to interact with Asn 429, an HOH molecule (which creates an h-bond bridge with Tyr 386), and an h-bond acceptor in the other part of the binding pocket to interact with the peptide backbone of Asp 377; (2) two aromatic groups to fill in the subpockets between Val 435, Leu 383, and between Pro 371, Leu 381; and (3) any atom group to fill in the subpocket among Tyr 428, Asn 429, and Leu 383. In contrast, in the right picture (5fe5), the model should contain slightly different binding properties: (1) an h-bond acceptor to interact with Asn 803, an HOH molecule (which creates an h-bond bridge with Tyr 760); (2) two aromatic groups to fill in the binding pocket and potentially create stacking interactions with Tyr 809.
Examples of molecular docking simulation to bromodomains
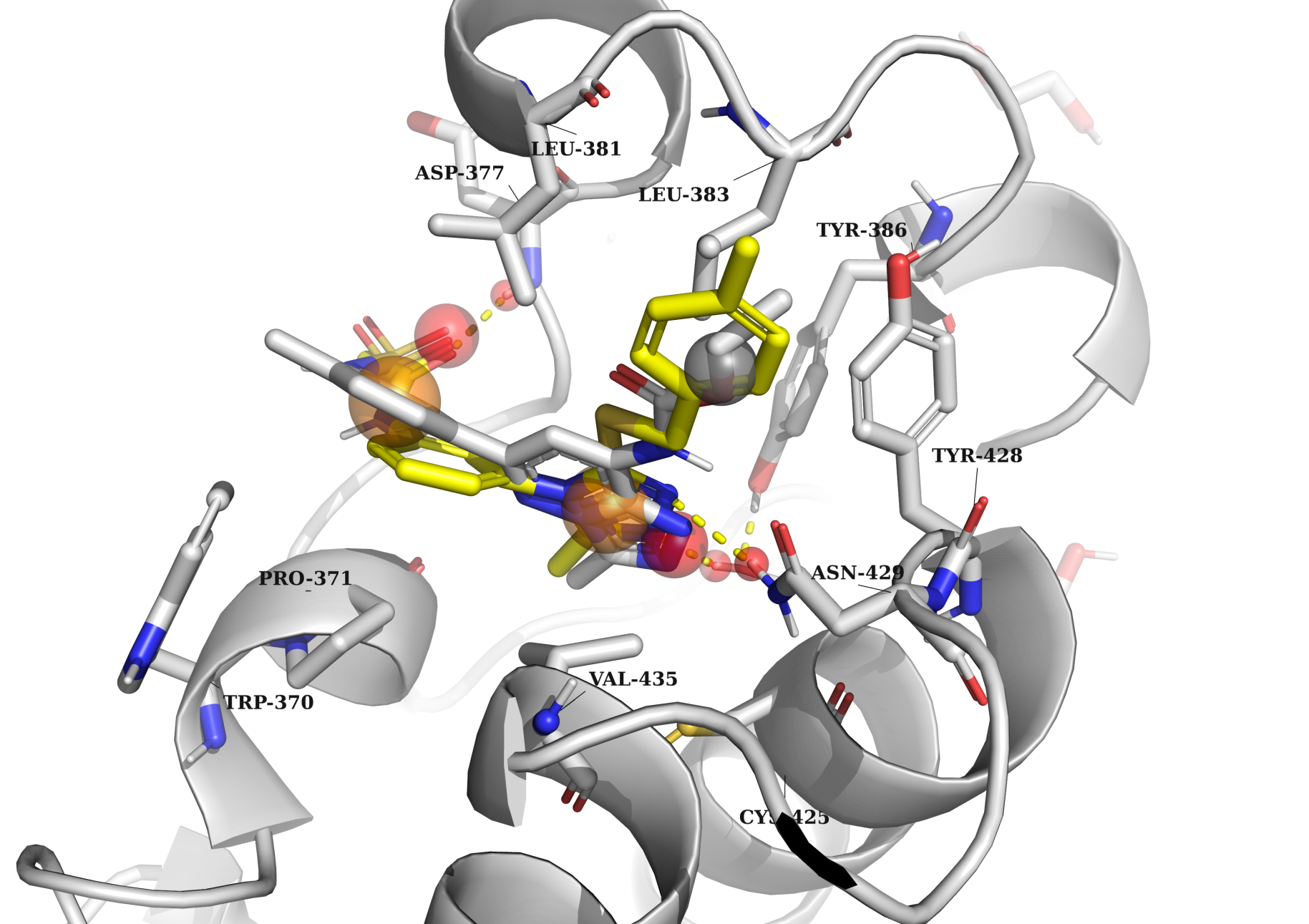
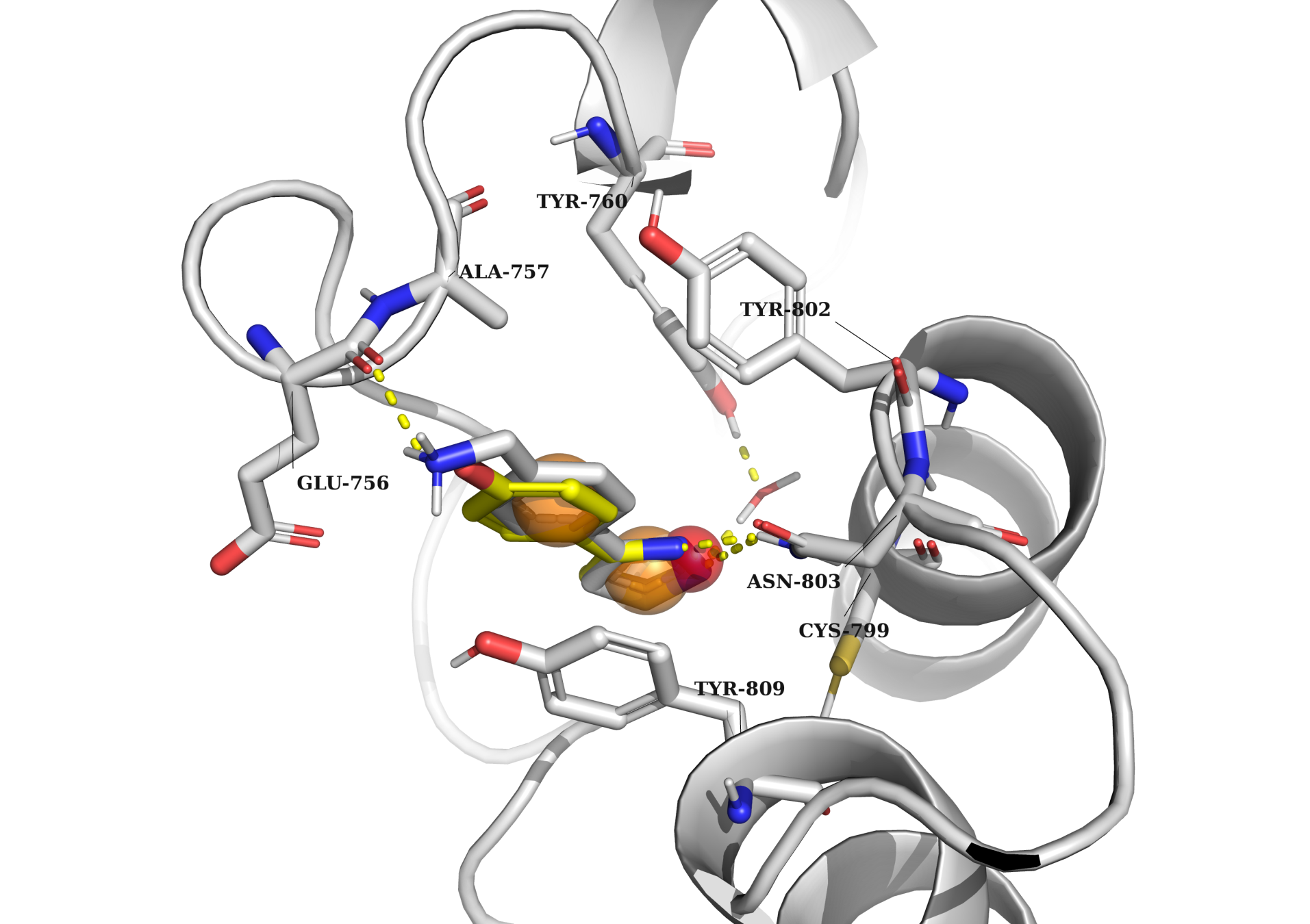
The molecular docking simulations to bromodomains were conducted using the following color scheme: red spheres represent h-bond acceptors, blue spheres represent h-bond donors, orange spheres represent aromatic groups, and grey spheres represent any other atom types. The protein and its native ligand are highlighted in grey, while an example of a docked ligand is shown in yellow.
Collection of referred small molecules with carefully collected activity data
2 405 compounds
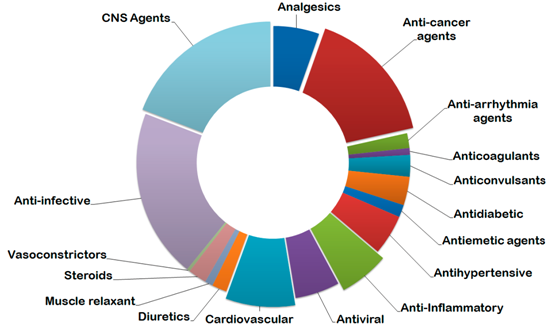
To address continuously growing interested to Drug Repurposing we designed and carefully collected a Bioactive Reference Collection of over 2 400 compounds with extensive target classes’ coverage and the broadest possible therapeutic areas applications – from CNS agents and anti-infectives to anticancer drugs and steroids.
Represented collection of carefully selected compounds includes 1 123 FDA approved drugs, as well as “tool compounds” with validated biological activity, active metabolites/prodrugs, and drug candidates that are currently undergoing clinical trials.
Download SD files
Library code: FAD-1123
Version: 9 November 2023
1 123 compounds
sublibrary of Bioreference Library
Library code: BAC-1282
Version: 14 November 2023
1 282 compounds
sublibrary of Bioreference Library
Ready-to-use, fully referred, alternative for compound screening.
Any compound from BRC collection can be ordered individually as focused sets: target-based or by therapeutic area.
Proven and reliable
- Stringent quality control using most advanced methods.
- Synthetic chemistry capacity with professionals in organic chemistry experienced in diverse synthetic methods and techniques.
- Drugs synthesis and their functional modifications for target identification and other purposes.
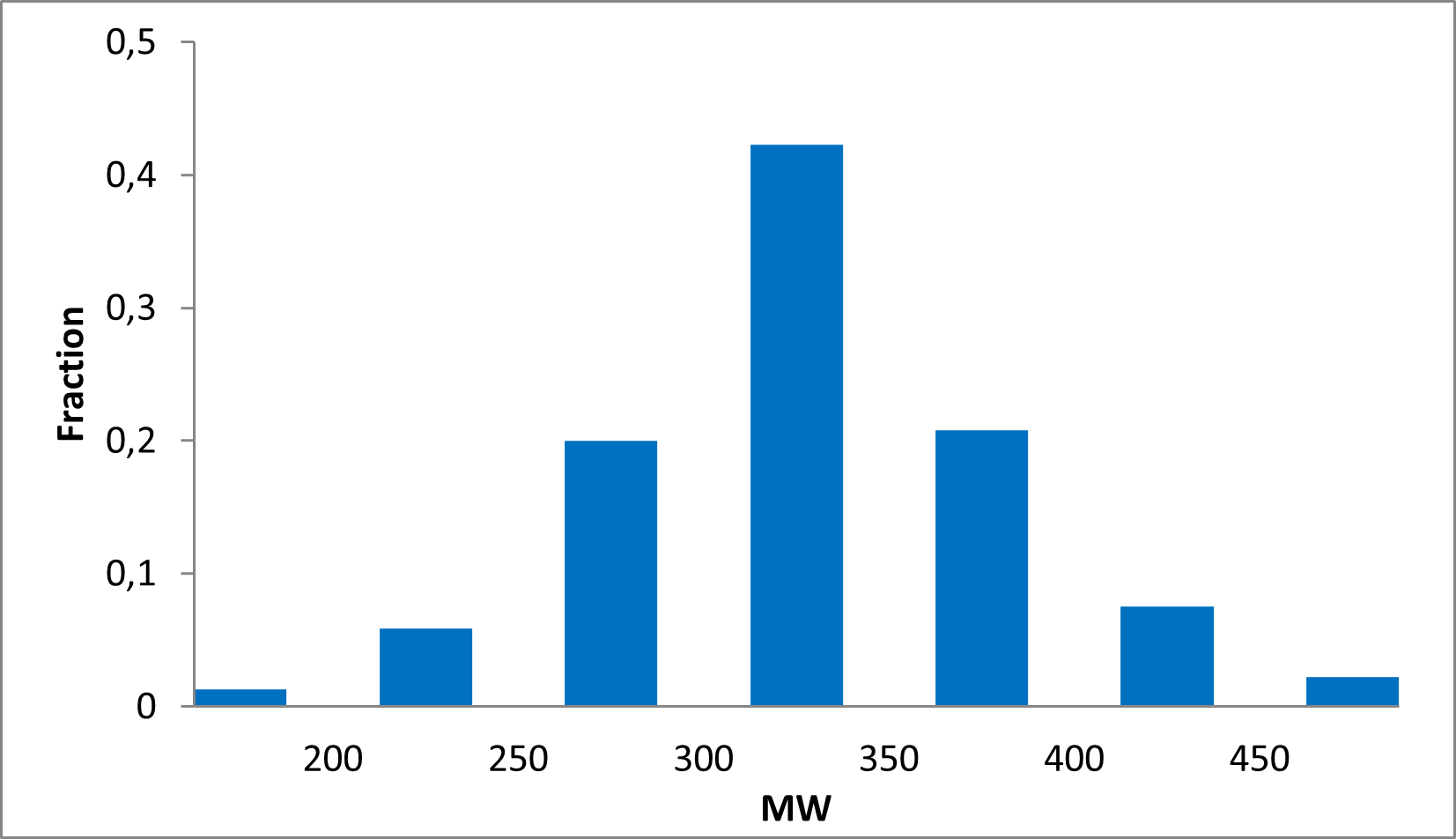
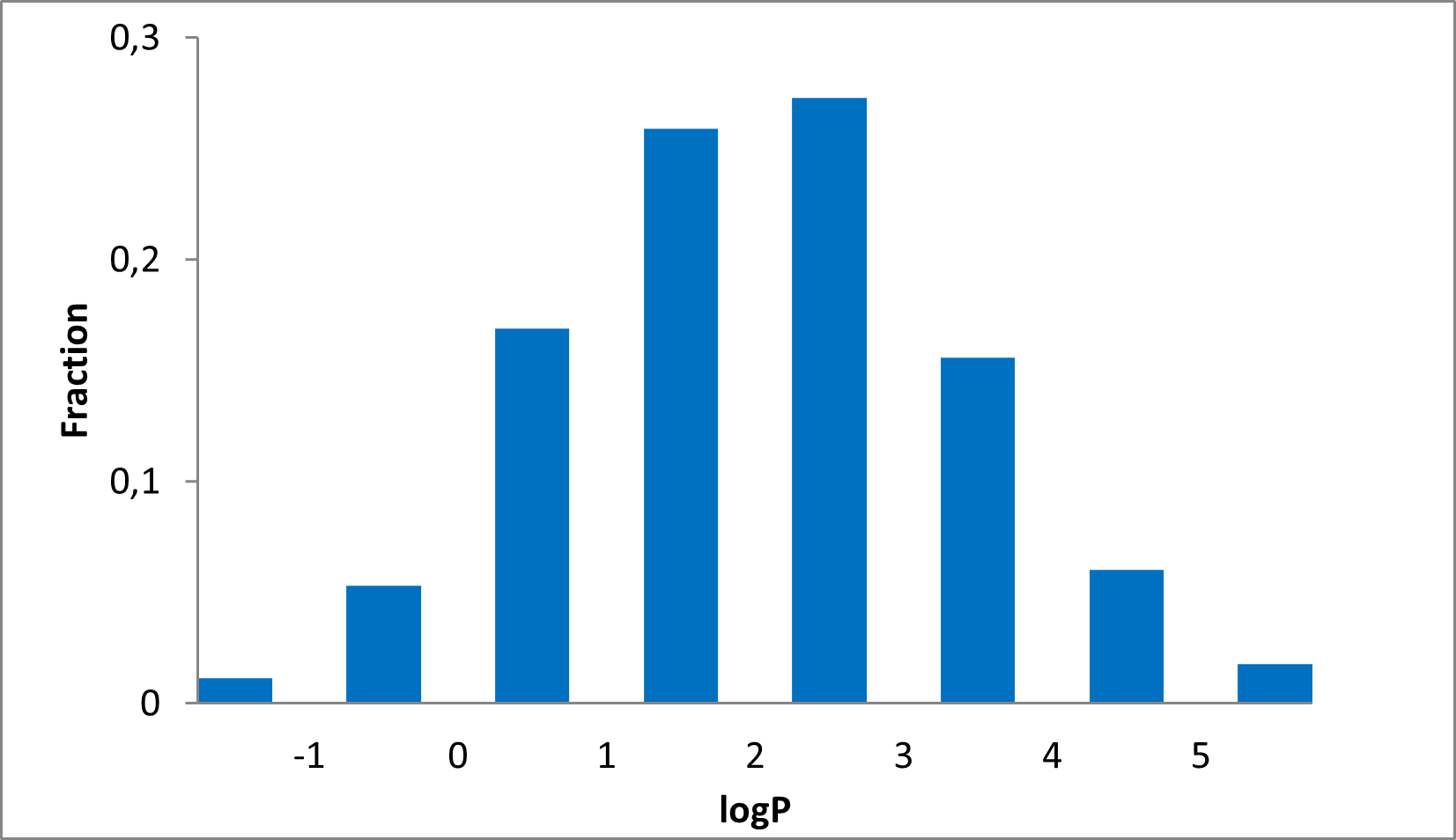
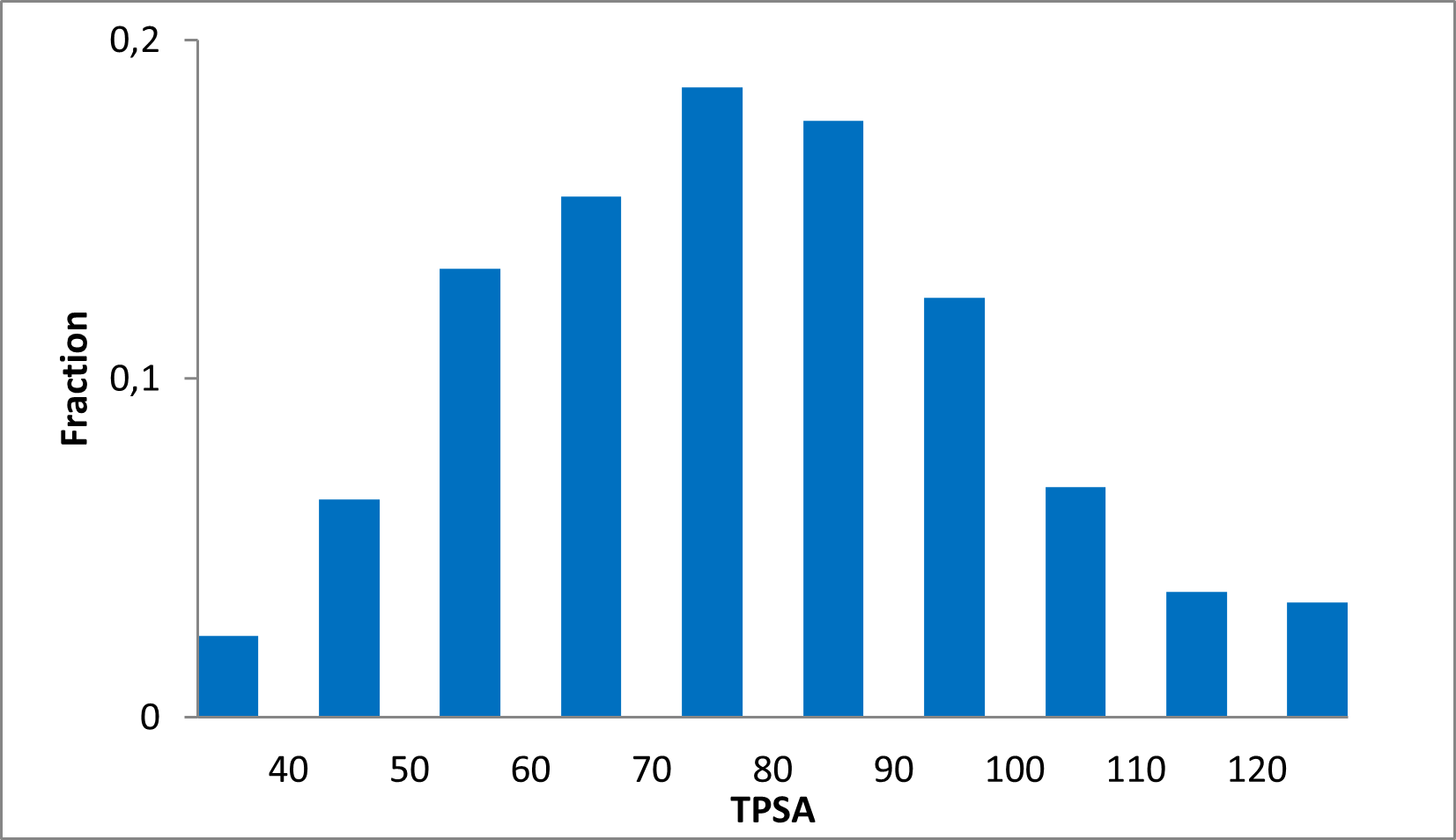
Distribution of compounds by therapeutic area
Related products & services
Target identification toolbox
Additionally we provide broad functionalization of Drugs and relative actives with covalent warheads, biotin or/and dye-linking, PED-derivatization and other modifications. Versatile chemistry and the largest stock of valuable reagents enables Enamine to produce new functionalization of well-known drugs.
We combined most diverse approaches and possible structural modifications to achieve most complete representation of our Target identification tools.
We offer over 100 ready-to-deliver pre-plated compound libraries in various custom formats. Our well-equipped liquid-handling department will make a library copy in any format convenient for your project.
Diversity Libraries
HLL-460
Size
460 160
compounds
Description
The largest diversity library with high MedChem tractability
Download file
HLL-200
Size
200 000
compounds
Description
Sublibrary of HLL-460
Download file
HLL-100
Size
100 160
compounds
Description
Sublibrary of HLL-460
Download file
DDS-50
Size
50 240
compounds
Description
Top-quality diverse library of recently synthesized compounds
Download file
DDS-10
Size
10 240
compounds
Description
High-quality diverse library of latest compounds
Download file
Enantioselective AS-MS Library
E-ASMS-12642
Size
12 642
compounds
Description
Innovatively designed and experimentally confirmed library for reliable enantioselective ASMS screening
Download file
CSL-11760
Size
11 760
compounds
Description
Diverse covalent library with most demanded warhead types
Download file
PSL-5760
Size
5 760
compounds
Description
Special diversity library created for Phenotypic Screens
Download file
PAINS-320
Size
320
compounds
Description
Special diverse selection of frequent hitters
Download file
Size
83
compounds
Description
Curated selection of frequent hitters
MCR-500
Size
500
compounds
Descriptions
Original design beyond Ro5 compounds
Download file
Covalent Libraries
CSL-11760
Size
11 760
compounds
Description
Diverse covalent library with most demanded warhead types
Download file
Covalent Serine Hydrolase Library
CSHL-12160
Size
12 160
compounds
Description
Designed for discovery of mild electrophilic inhibitors of the largest enzyme class
Download file
Coronavirus Mpro covalent Library
MPC-2640
Size
2 640
compounds
Description
Designed for the discovery of new SARS-CoV-2 and pan-Coronavirus antivirals
Download file
CFL-8480
Size
8 480
compounds
Description
Diverse covalent warheads with balanced reactivity
Download file
Cysteine-Focused Covalent Library
CYS-3200
Size
3 200
compounds
Description
Library of Cys-specific covalent electrophilic binders
Download file
Serine-Focused Covalent Library
SER-1600
Size
1 600
compounds
Description
Special selection of Serine focused irreversible binders
Download file
Lysine-Focused Covalent Library
LYS-1600
Size
1 600
compounds
Description
The ultimate selection of Lys-specific binders
Download file
Electrophilic Covalent Probe Library
ECPL-960
Size
960
compounds
Description
Characterized by a new HTS thiol-reactivity assay
Download file
sACR-4080
Size
4 080
compounds
Descriptions
Diverse screening Acrylamides pre-plated at 10 mM concentration
Download file
fACR-2240
Size
2 240
compounds
Descriptions
Representative selection of fragment Acrylamides pre-plated at 100 mM stock concentration
Download file
sCLA-1200
Size
1 200
compounds
Descriptions
Library of diverse HTS-size chloroacetamides pre-plated at 10 mM concentration
Download file
Chloroacetamide Fragment Library
fCLA-1360
Size
1 360
compounds
Descriptions
Diverse strict Ro3 compliant chloroacetamides plated at 100 mM stock concentartion
Download file
SFF-640
Size
1 120
compounds
Descriptions
Representative selection of N-, O-linked and Aryl sulfonyl fluorides within fragment space
Download file
Size
960
compounds
Description
Enantiomeric pairs of covalent electrophilic fragments
Download file
CMF‑141
Size
141
compounds
Description
Covalent Heterocyclic Fragment Library for identification of Cryptic and Allosteric Pocket
Download file
Molecular Glue
DCAF-5440
Size
5 440
compounds
Descriptions
Designed to target DCAF family of E3 ligases
Download file
IMiD Library
IMiD-4900
Size
4 900
compounds
Descriptions
Designed and specially synthesized for the discovery of Immunomodulatory Imide Drugs
Download file
CRBN-MG-4560
Size
4 560
compounds
Description
Designed to capture the widest possible range of novel chemotypes of CRBN binders, providing a foundation for the discovery of next-generation Molecular Glues
Download file
PAG-640
Size
640
compounds
Description
Built upon novel Phenyl Amino Glutarimide (PAG) analogs, these compounds combine structural innovation with potent activity — offering a fresh perspective in Molecular Glue design
Download file
PD-880
Size
880
compounds
Description
Chemical stability, cellular potency, and permeability are just a few of the advantages that make Phenyl Dihydrouracil (PD)-based CRBN ligands promising candidates for optimal Molecular Glue discovery
Download file
PG-800
Size
800
compounds
Description
Phenyl Glutarimide (PG)-based library was purposefully developed to expand the structural diversity space of Molecular Glues
Download file
Len Library
Len-640
Size
640
compounds
Description
Lenalidomide- and Pomalidomide-scaffold based — a time-tested backbone, reimagined for modern Molecular Glue design
Download file
AAG Library
AAG-1600
Size
1 600
compounds
Description
Acylated Amino Glutarimide (AAG) featuring 5- and 6-membered heterocyclic systems: a promising platform for the discovery of next-generation Molecular Glues
Download file
Avadomide library
AVD-560
Size
560
compounds
Descriptions
A special selection of new Avadomide analogs
Download file
Diverse CRBN Library
CRBN-960
Size
960
compounds
Descriptions
Designed to cover all structural diversity of CRBN binders, a key component of the E3 ubiquitin ligase complex
Download file
CRBN Covalent Library
CCRBN-160
Size
160
compounds
Descriptions
A small selection of diverse covalent binders capable of interacting with CRBN
Download file
VHL ligands and intermediates
VHL-80
Size
80
compounds
Descriptions
A versatile tool for discovering new VHL-based degraders
Download file
AI-enabled Libraries
Size
1 600
compounds
Description
Focused on targeting the top CRLs that have been explored as the most promising targets
Download file
Size
1 520
compounds
Description
Designed to target 3 HECT E3 Ligases for the discovery of new-generation drugs
Download file
Size
1 520
compounds
Description
Designed for discovery of RNF216, RNF19A, PRKN, RNF13 potent binders and modulators
Download file
AI-enabled USP Library
Size
1 200
compounds
Description
Designed for modulation of the largest DUBs family and delivery of a promising treatment for incurable diseases
Download file
AI-enabled GPCR Library
Size
1 760
compounds
Description
Designed to deliver new and efficient modulators of CCR5, HTR2A, HTR2B, MRGPRX1, CXCR6, CMKLR2, CCR10, GPR3, GPR4, GPR39, CCR4 receptors
Download file
HIPPO Pathway Library
Size
1 600
compounds
Description
Designed to deliver reliable hits for over 10 essential targets involved in the Hippo pathway
Download file
AI-enabled Molecular Chaperones Library
Size
1 360
compounds
Description
Designed for hit finding for Hsp90, Hsp70, Hsp60, HspD1, ClpP, Ch60 and Hsp100 protein targets
Download file
AI-enabled Allosteric Ion Channel Library
Size
2 080
compounds
Description
A new approach for modulating the most investigated and effective drug targets with good clinical records
Download file
AI-enabled PARP Library
Size
1 440
compounds
Description
Designed to target the most investigated and validated for drug discovery PARPs
Download file
AI-enabled TF Library
Size
1 520
compounds
Description
The hottest protein targets for discovering new treatments for the most challenging and not yet treated diseases
Download file
Targeted Libraries
AGR-10
Size
10 240
compounds
Description
Library of compounds intended for use in agro/crop science
Download file
AGR-14
Size
14 160
compounds
Description
Designed for discovery of novel allosteric ligands
Download file
Size
4 800
compounds
Description
Carefully selected molecules via docking and visual evaluation
Download file
ABAC-32
Size
32 000
compounds
Description
Designed for the discovery of novel antibacterials
Download file
ATB-2500
Size
2 500
compounds
Description
Designed for the discovery of new effective and safe treatment
Download file
AVR-3200
Size
3 200
compounds
Description
Designed for discovery of new Nucleoside-like antivirals
Download file
Size
1 348
compounds
Description
Designed for discovery of new water channels modifiers
Download file
Size
7 171
compounds
Description
Designed for discovery of novel BACE inhibitors
Download file
BRD-15
Size
15 360
compounds
Description
Specially selected molecules to target bromodomains
Download file
CICL-10560
Size
10 560
compounds
Description
Designed for discovery of new Voltage-gated calcium channel blockers
Download file
CNS-47
Size
47 360
compounds
Description
Library of novel small molecules with high CNS MPO scores
Download file
CNSd-5
Size
5 440
compounds
Description
Sublibrary of CNS-47 Library
Download file
COV-16800
Size
16 800
compounds
Description
Designed for the discovery of new SARS-CoV-2 and pan-Coronavirus antivirals
Download file
DNA-5760
Size
5 760
compounds
Description
Designed for identification of new actives against proteins essential for DNA stability
Download file
EPG-38080
Size
38 080
compounds
Description
Library of compounds focusing to hit on a number of epigenetic targets
Download file
ERL-8960
Size
8 960
compounds
Description
Designed to effectively target the receptor and block estrogen release
Download file
GML-2470
Size
2 470
compounds
Description
Specially synthesized set of compounds able to mimic glycosides and their interaction with proteins
Download file
GPR-53
Size
53 440
compounds
Description
Designed for discovery of new GPCR ligands
Download file
HBL-24
Size
24 000
compounds
Description
Designed for discovery of novel kinase ATP pocket binders
Download file
IDO-4800
Size
4 800
compounds
Description
IDO focused library designed by a combination of structure- and ligand-based methods
Download file
Size
45 760
compounds
Description
Designed for discovery of novel hits in Immuno-Oncology therapeutic area
Download file
ICL-36
Size
36 800
compounds
Description
Designed for discovery of new Ion Channels ligands
Download file
JAK-STAT-1280
Size
1 280
compounds
Description
Designed for efficient hit finding against a number of immune disorders, including RA
Download file
KNS-64960
Size
64 960
compounds
Description
Designed for discovery of novel protein kinase inhibitors
Download file
KYN-13
Size
13 120
compounds
Description
Designed for discovery of new regulators of methabolic disorders
Download file
LGR-6400
Size
6 400
compounds
Description
A sub-library of Enamine’s GPCR Library designed for discovery of novel lipid GPCR ligands
Download file
Size
1 388
compounds
Description
A set of LOXs inhibitors designed using docking and 2D similarity search
Download file
Size
2 468
compounds
Description
A set of compounds focused on targeting molecular chaperones
Download file
NML-320
Size
320
compounds
Descriptions
Small library of specially synthesized compounds
Download file
PDZ Domain Library
PDZ-1920
Size
1 920
compounds
Descriptions
Sublibrary of PPI-40
Download file
PML-8960
Size
8 960
compounds
Descriptions
Selected molecules able to mimic common protein motifs
Download file
Size
40 640
compounds
Descriptions
Designed for discovery of novel PPI inhibitors
Download file
RNA-28
Size
28 000
compounds
Descriptions
Designed to promote the discovery of new-generation medicines
Download file
CSHL-12160
Size
12 160
compounds
Descriptions
Designed for discovery of mild electrophilic inhibitors of the largest enzyme class
Download file
SICL-5440
Size
5 440
compounds
Descriptions
Designed for discovery of new Nav1.7 channel blockers
Download file
TBL-3200
Size
3 200
compounds
Description
Library of potential tubulins ligands
Download file
WPL-10
Size
10 560
compounds
Description
Designed for the discovery of new effective modulators of Wnt/β-catenin signaling pathway
Download file
Fragment Libraries
ESS-320
Size
320
compounds
Description
Elaborated tool for initial screen
Download file
High Fidelity Fragment Library
HFF-1920
Size
1 920
compounds
Description
Fragments of high MedChem tractability
Download file
DSI-860
Size
860
compounds
Description
Designed for easy and rapid follow-up synthesis
Download file
MiniFrags-80
Size
80
compounds
Description
Guiding optimisation of fragment-derived lead compounds
Download file
CFL-8480
Size
8 480
compounds
Description
Diverse covalent warheads with balanced reactivity
Download file
FDS-1000
Size
1 000
compounds
Description
Specially designed for 19F NMR ligand-based screening
Download file
Size
800
compounds
Description
Designed for easy and efficient exploration of novel protein targets
Download file
Natural Product-like Fragments
NPL-4160
Size
4 160
compounds
Description
Source of biologically validated starting points
Download file
3D Shape Diverse Fragment Library
3DF-1200
Size
1 200
compounds
Description
Unique 3D diversity among shaped molecules
Download file
PPIF-3600
Size
3 600
compounds
Description
Fragments able to mimic protein structural motifs and hot-spot residues
Download file
Single Pharmacophore Fragments
SPF-1500
Size
1 500
compounds
Description
Fragments for easy-to-analyse protein-ligand interaction
Download file
Carboxylic Acid Fragment Library
CAF-4000
Size
4 000
compounds
Description
Designed for specific protein targets and sensible onset
Download file
Size
1 280
compounds
Description
The most medchem reliable source of carboxylic acids replacement
Download file
Halogen-enriched Fragment Library
HEF-1920
Size
1 920
compounds
Description
Library of high diversity of halogen bonding motifs
Download file
Electrophilic Covalent Probe Library
ECPL-960
Size
960
compounds
Description
Characterized by a new HTS thiol-reactivity assay
Download file
Covalent Heterocyclic Fragment Library
CovHetLib‑141
Size
141
compounds
Description
Covalent Heterocyclic Fragment Library for identification of Cryptic and Allosteric Pocket
Download file
CNSF-1
Size
1 280
compounds
Description
CNS-friendly molecules capable of BBB penetration
Download file
Size
960
compounds
Description
Enantiomeric pairs of covalent electrophilic fragments
Download file
Size
372
compounds
Description
Size-optimized fragment library efficiently covering pharmacophore space, developed using ChemPass's Universal Fragment Library (UFL) Design Platform.
Download file
Size
2 301
compounds
Description
General fragment library developed with ChemPass's Universal Fragment Library (UFL) Design Platform.
Download file
Bioactive Libraries
CLOUD-299
Size
299
compounds
Description
Represents the diversity of structures and molecular targets of all FDA-approved chemical entities (Nat Chem Biol 13 (2017), 771–778)
BRL-2405
Size
2 405
compounds
Description
Actives with extensive target classes’ coverage and the broadest possible therapeutic areas applications – from CNS agents and anti-infectives to anticancer drugs, steroids and molecular glues.
FAD-1123
Size
1 123
compounds
Description
Most relevant selection of drugs in one formulation/no duplicates. Contains WHO List of Essential Medicines including Lapatinib and latest approved drugs such as Tivozanib, etc.
Bioactive sublibrary
BAC-1282
Size
1 282
compounds
Description
Compounds with referred biological activity for repurposing or investigation of new pathways and mechanisms
PSL-5760
Size
5 760
compounds
Description
Library of cell penetrated compounds and their closest analogs. Covers diverse therapeutic areas from antitumor, neurology and antibacterial to aging diseases.
PAINS-320
Size
320
compounds
Description
Frequent hitters with most diverse scaffold selection – from small hydroquinone and other covalent modifiers to polyfluorinated highly lipophilic molecules and dyes.
Size
83
compounds
Description
Curated selection of frequent hitters
Focused Sets
Special diverse selection of frequent hitters
320 compounds
Pan-Assay Interference Compounds (PAINS) are the most recognized filters among medicinal chemists. Since first publication in 2010 by John Bell these filters have become industry standard in Drug Discovery. While carefully removing all PAINS-related compounds from Enamine libraries we realized that these compounds can be very useful in HTS assay set-up & validation.
Special diversity set of PAINS available for prompt delivery in various plated formats including the most popular listed below:
Typical Formats
PAINS Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
PAINS-320-0-Z-10
Compounds
320
1 plate
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
PAINS-320-10-Y-10
Compounds
320
1 plate
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
PAINS-320-50-Y-10
Compounds
320
1 plate
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
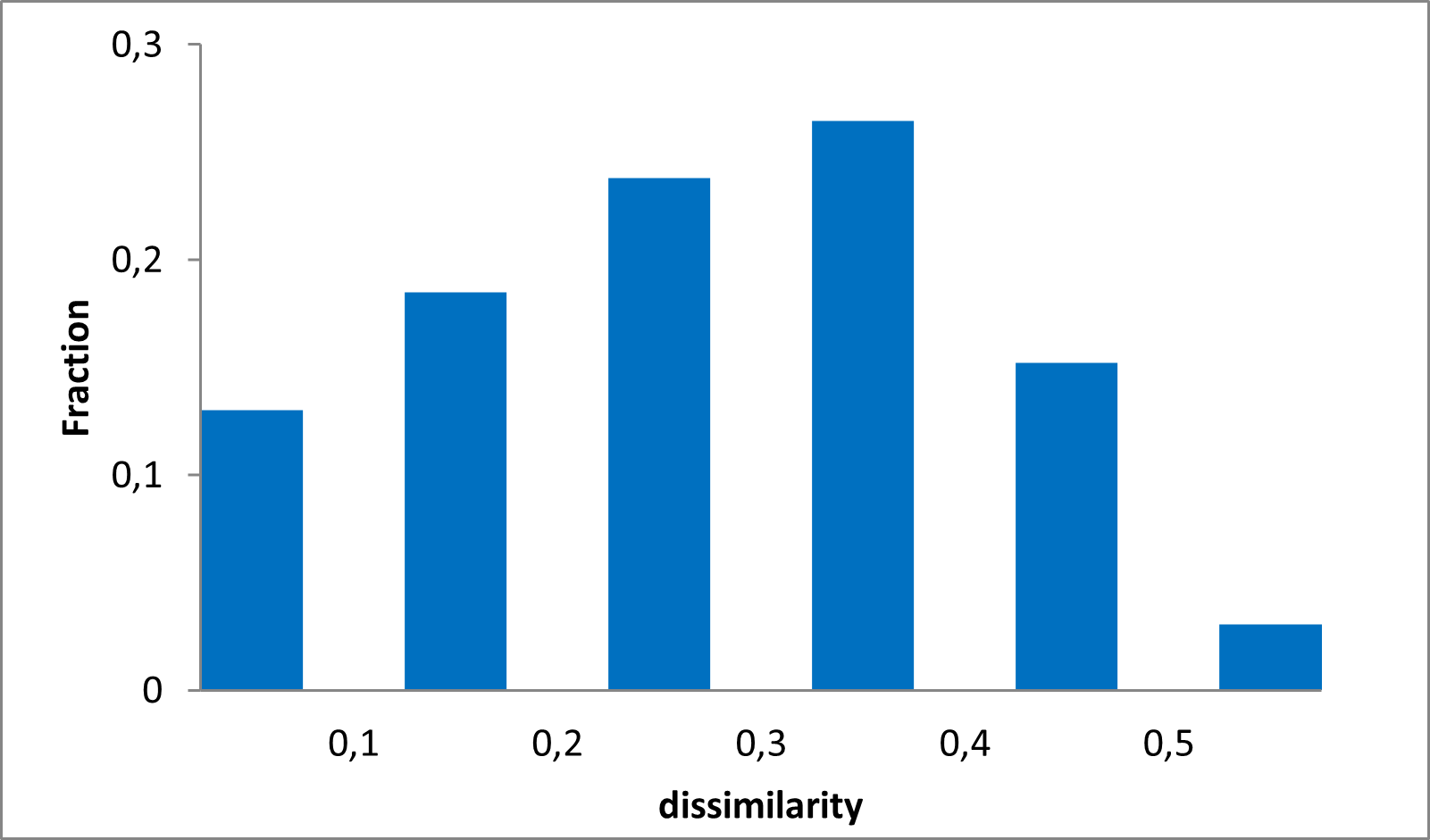
Library design
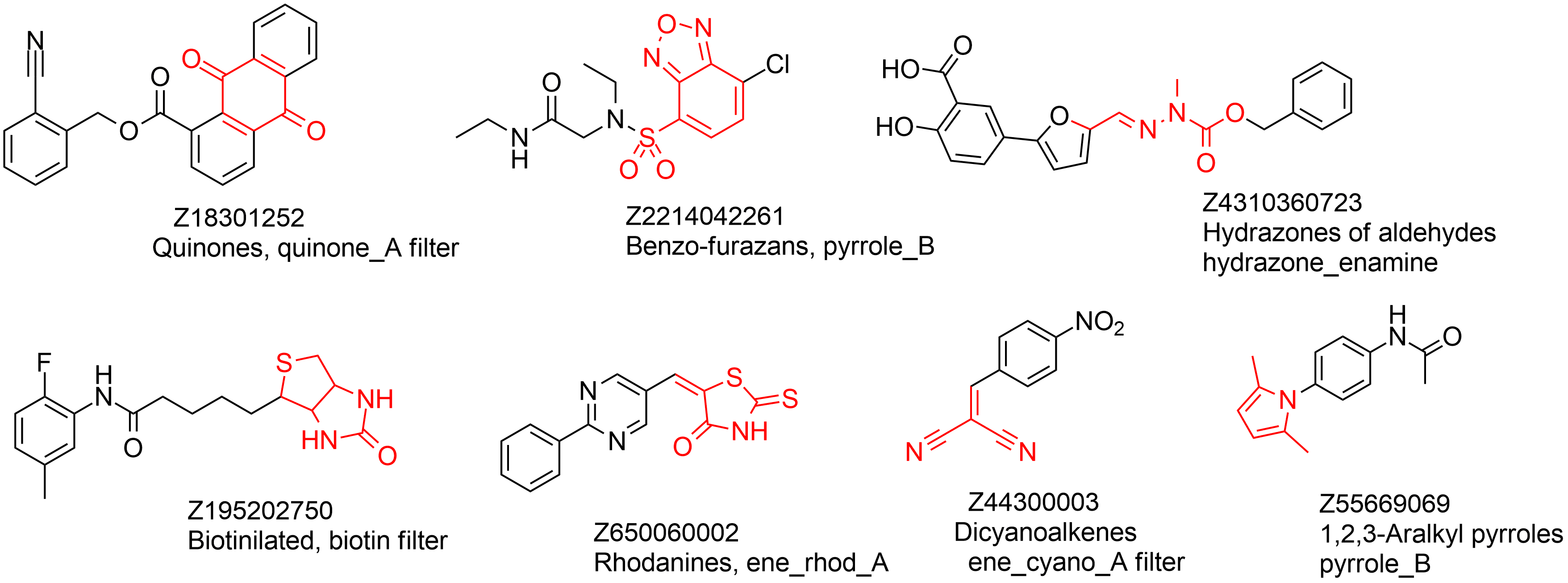
The design is based on substructural motifs of known PAINS compounds. We used originally reported PAINS filters to identify a set of over 80k in-stock compounds. This set was clustered using fingerprint-based approach and Tanimoto similarity distance calculations. The most populated clusters with at least 5 compounds were extracted and one representative example was selected from each cluster resulting in a library of 320 compounds. The library of most diverse 320 PAINS is available in plate format for your convenience. Examples are given below.
Key features
- Represents the most common false positives
- Substructure & scaffold diversity
- All compounds suitable for storage as DMSO solutions
Example of structures and their specific PAINS filters