Designed for discovery of novel kinase ATP pocket binders
24 000 compounds
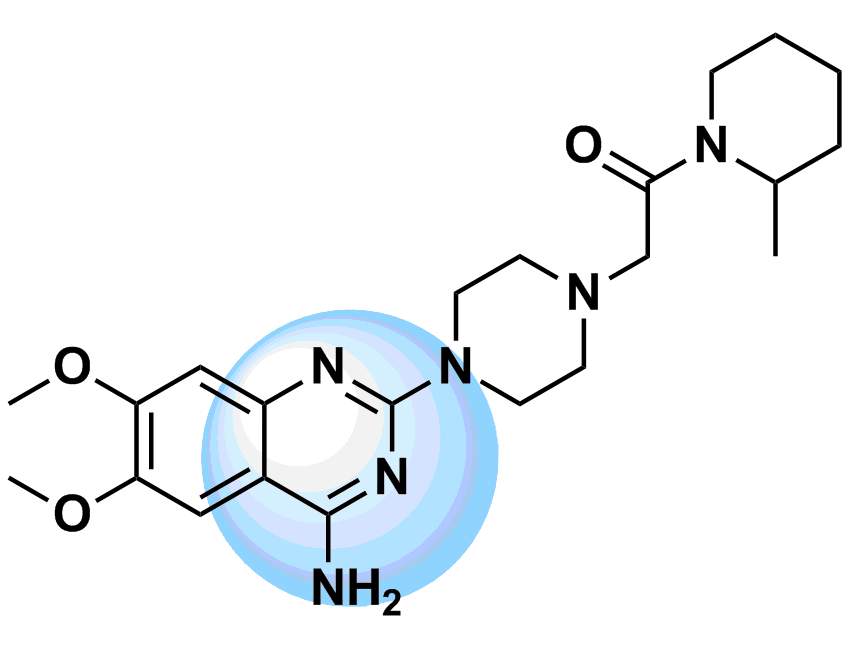
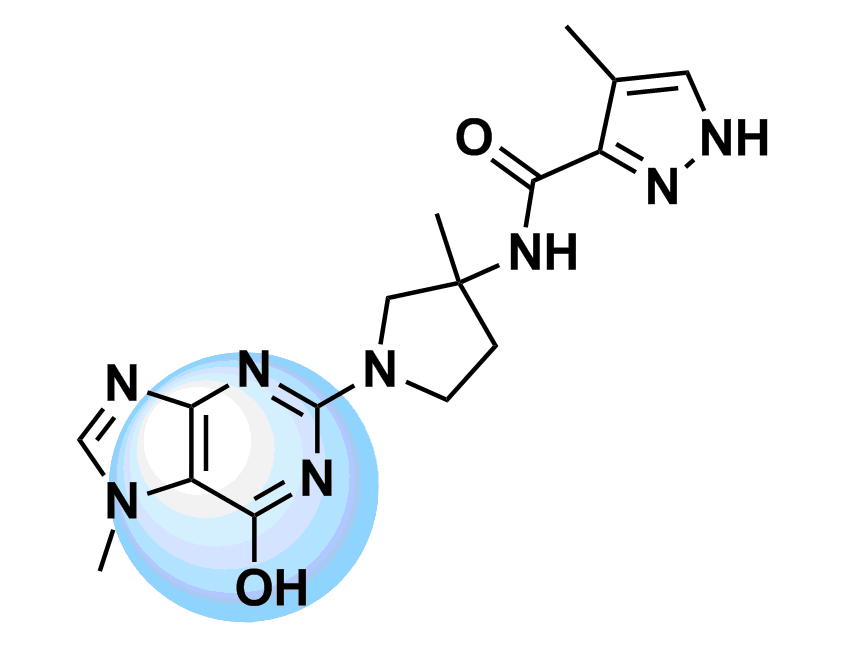
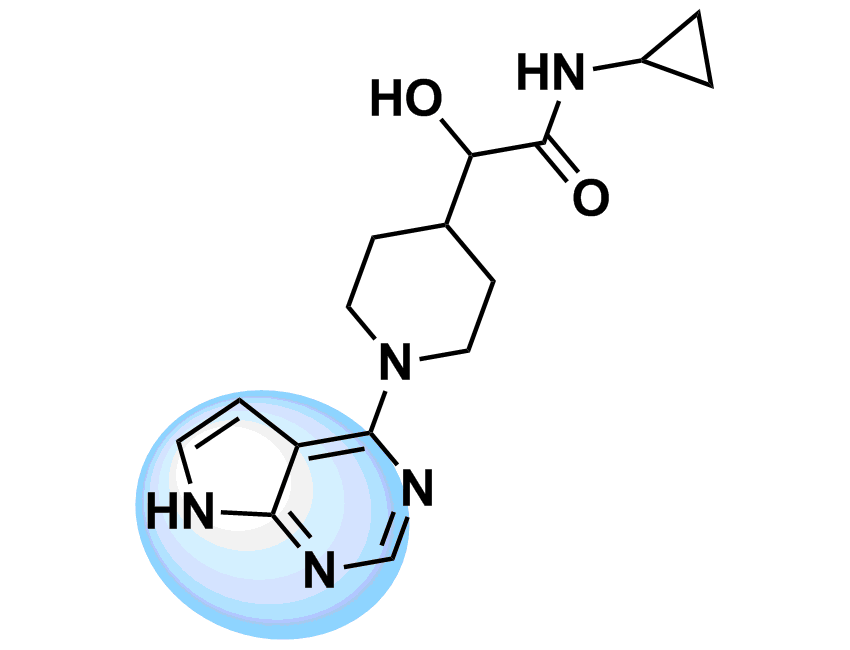
The most straightforward approach to the design of kinase inhibitors relies on targeting ATP pocket. In fact, all compounds of this class approved by FDA (Copanlisib, Neratinib, Brigatinib, and Ribociclib) demonstrate this binding mode. The standard kinase interaction pattern consists of a hydrogen bond acceptor for the hinge region, heteroaromatic core with various substituents, and a second hydrogen acceptor for the conserved Lysine residue. This type of Inhibitors can be considered as ATP mimetics in the sense that they make interactions similar to that of ATP. Hydrogen bonds with Hinge Region formed by the adenosine moiety of ATP are crucial for effective binding (Figure 1, left). Analysis of a broad number of the kinase-inhibitor interactions has shown that similar hydrogen bonding patter is necessary for high inhibitory potency.
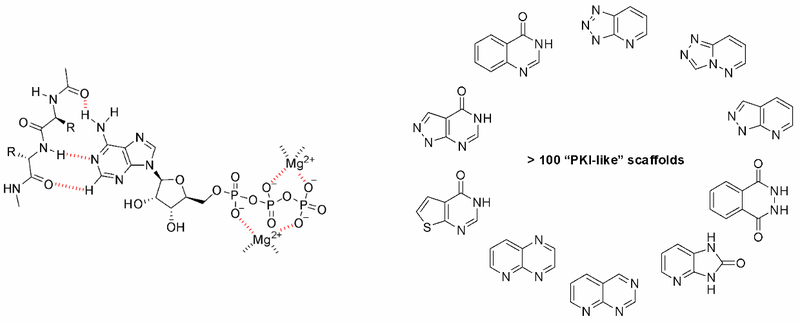
Fig. 1. ATP binding mode with hinge region (left) and examples of core structures that can mimic that mode (right).
Typical Formats
Hinge Binders Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
HBL-24-0-Z-10
Compounds
24 000
19 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
HBL-24-10-Y-10
Compounds
24 000
75 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
HBL-24-50-Y-10
Compounds
24 000
75 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
HBL-24-10-Y-10 screening library 24 000 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library code: HBL-24
Version: 16 March 2021
24 000 compounds
sublibrary of KNS-65
Library design
Starting from the results of our previous study (Anticancer Agents Med. Chem. 2007, 7, 171–188) and detailed structure analysis of known and most potent kinase inhibitors, we have developed a series of unique structure filters aimed to identify potential inhibitors targeting ATP pocket. Molecular fragments able to form at least two hydrogen bonds with hinge region (Figure 1, right) have been analyzed, and appropriate topological models were developed for searching of directed inhibitors. The screening models were evaluated with a reference set of known kinase inhibitors (over 2 000 molecules with high inhibitory activity). Validated topological models were applied to over 3.2 M Enamine stock collection to produce Kinase Hinge Binders Libarary. The resulting set of compounds was evaluated with main emphasis on novel chemotypes. MedChem filters including PAINS and Ro5 PhysChem restrictions were applied as a prerequisite.