Carefully selected molecules via docking and visual evaluation
4 800 compounds
Development of efficient kinase inhibitors has been a long-standing challenge in drug development. The structural and mechanistic characterization of kinase inhibitor provides new strategies to develop specific kinase inhibitors by targeting a binding pocket adjacent or not adjacent to the ATP binding pocket.
Allosteric kinase inhibition is among the most promising and sensitive deactivation mechanism of kinase activity. Four type of possible allosteric kinase pockets were determined by now:
- Myristoyl pocket;
- Inhibitor binding mode that occupies part of binding pocket adjacent to the ATP-site;
- PIF-pocket (regulatory site targeted);
- Pocket type I1/2 relatively allosteric regulators of kinases.
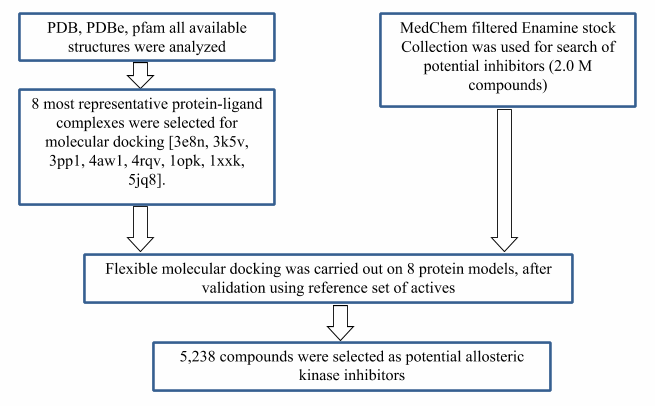
All available PDB structures with kinase allosteric inhibitors were analyzed. Most representative structures were used for molecular docking calculation. General algorithm of library design represented on the scheme below:
Typical Formats
Allosteric Kinase Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
ALK-4-0-Z-10
Compounds
4 800
4 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
ALK-4-10-Y-10
Compounds
4 800
15 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
ALK-4-50-Y-10
Compounds
4 800
15 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
ALK-4-10-Y-10 screening library 4 800 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD file
Library code: ALK-4
Version: 17 February 2022
4 800 compounds
sublibrary of KNS-64
Examples of the molecules

Fig. 1. Example of molecular docking result into the Myristoyl pocket (4aw1 PDB entry). Native ligand represented in grey sticks, docked ligand – in yellow.
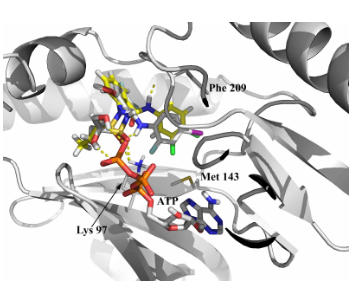
Fig. 2. Example of molecular docking into the ATP-kinase subpocket (3e8n PDB entry). Docking calculations were performed with CoA features.
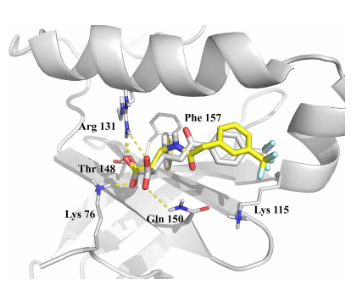
Fig. 3. Example of molecular docking into the PIF pocket (4aw1 PDB entry).
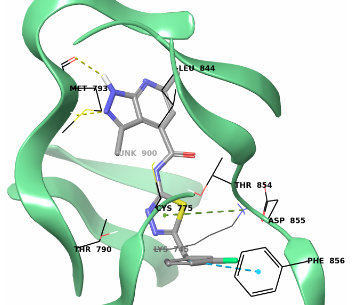
Fig. 4. Example of molecular docking into the PIF I1/2 binding site (4aw1). Hit binding mode.



