Fragments able to mimic protein structural motifs and hot-spot residues
3 600 compounds
Protein-protein interactions regulate most aspects of life cycle thereof being the most attractive and perspective target for contemporary drug development. This field is still not well explored and there are no common rules for proteins involved in this type of interaction. Such intricate biological systems cannot be cost-efficiently tackled using conventional high-throughput screening methods and algorithms. However, fragment-based approach showed fruitful results in search of new PPI inhibitors. We created new library of PPI fragments with dedicated design that consists of systematical knowledge of common PPI inhibitors and selection by privileged structural motifs.
Using our PPI Fragments for hit finding you receive multiple benefits allowing you to save on time and costs in lead generation. Each fragment from the library can be easily followed with available in stock analogues or synthesis of new derivatives through our REAL Database technology.
Typical Formats
Catalog No.
PPIF-3600-Y-10
Compounds
3 600
10 plates
Amount
10 µL of 10 mM DMSO-d6 stock solutions
Plates and formats
384-well microplates, Echo qualified Labcyte LP0200
Price
Catalog No.
PPIF-3600-X-50
Compounds
3 600
40 plates
Amount
50 µL of 10 mM DMSO-d6 stock solutions
Plates and formats
96-well plates, Greiner, first and last columns empty, 80 compounds per plate
Price
Catalog No.
PPIF-3600-X-100
Compounds
3 600
40 plates
Amount
100 µL of 10 mM DMSO-d6 stock solutions
Plates and formats
96-well microplates, first and last columns empty, 80 compounds per plate
Price
Catalog No.
PPIF-3600
Compounds
3 600
Amount
Custom
Plates and formats
Any custom format
Price
Download SD file
Library code: PPIF-3600
Version: 16 September 2021
3 600 compounds
at 40 mM in DMSO-d6
Key features
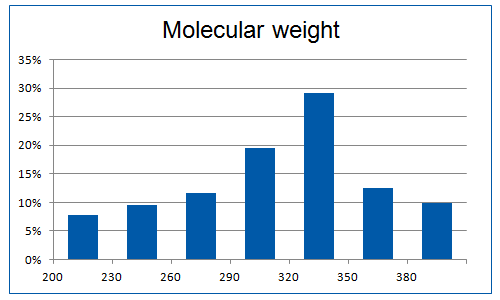
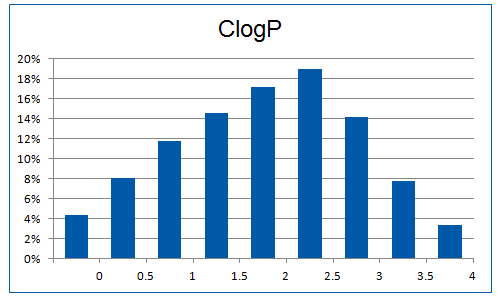
Recent researches in the field of PPI fragments revealed the requirement of higher molecular weight molecules compared to common fragments due to a larger contact surface area. Therefore “Rule of four” restriction were used to extract initial PPI fragments subset, as they are typically larger and more lipophilic. Additionally, machine-learning method (decision tree) with number of validated descriptors was used to refine the library.
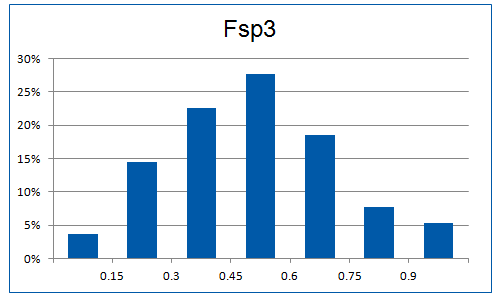
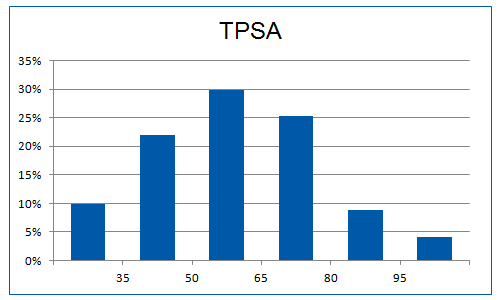
The “hot-spots” concept has been applied, implying the use of “key” amino acid residues involved in PPIs. A significant number of the selected fragments contains groups/moieties which correspond to these hot-spots. Additionally, library was enriched with the molecules having alpha helix-like structure, able to mimic protein motifs. Also since hydrogen bonds often play a crucial role in PPIs the preference was set for the molecules bearing at least one H-bond donor (>70%) and one H-bond acceptor (100%).