Designed for the discovery of new effective and safe treatment
2 500 compounds
Tuberculosis (TB) is among the most threatening diseases that can cause the next pandemic across the world. It is one of the most rampant infectious diseases owing largely to the methods of transmission and high death ratio. The absence of efficient treatment and the rapid appearance of drug resistance make TB one of the most emerging tasks to resolve in public health. Continuously mutated, Mtb remains one of the most dangerous and difficult-to-treat bacterial diseases. Lack of early diagnosis and the prevalence of resistant strains make TB extremely difficult for timely detection and treatment. Antituberculosis drug development is a high priority to prevent future pandemics. It needs further investment in scientific research and rational drug design to combat the high death rate caused by this disease.
The library is available in pre-plated formats for quick access and is supported with multiple benefits on hit follow-up studies:
- Analogs and hit samples resupply from dry stock of over 4.4 M compounds with immediate QC check.
- MedChem support in hit follow-up and optimization to lead series.
- Same-day ADME tests for design and synthesis prioritization.
Typical Formats
Catalog No.
ATB-2500-10-Y-10
Compounds
2 500
14 plates
Amount
10 µL of 10 mM DMSO solutions
Plates and formats
384-well plates Greiner #781280, 320 compounds per plate, first two and last two columns empty
Price
Catalog No.
ATB-2500-50-X-10
Compounds
2 500
53 plates
Amount
50 µL of 10 mM DMSO solutions
Plates and formats
96-well plates, Greiner #650160, 80 compounds per plate, 1 & 12 columns empty
Price
Catalog No.
Library & follow-up package
Plates and formats
ATB-2500-10-Y-10 library, hit resupply from dry stock and fresh DMSO solutions plus 240+ analogs from stock and follow-up synthesis from REAL Space
Price
*We will happily provide our library in any other most convenient format for your project. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or, Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400) or send your preferred labware. Compounds pooling can be provided upon request.
Download SD file
Library Design
To design the library and select the most promising molecules, we pay attention to the complexity of Mtb cell-wall and the difficulties of membrane penetration. Despite the absence of certain ruleless and defined filters, we tried to remove all compounds that could have issues with penetration based on the reported data and structure analysis of cell active vs inactive compounds.
Having studied some special features related to the biochemical pathways and morphological properties inherent to Mtb, a group of protein targets has been selected and prioritized to perform in silico screening (vHTS) and select compounds with the alleged activity. Generally, the protein targets in this library can be divided into two subsets: Cell Wall Synthesis-associated proteins and Mtb-specific Essential protein targets distinct from those present in eukaryotic organisms.
Alanine racemase (Alr), which catalyzes the interconversion of L-alanine and D-alanine. Which in turn plays a key role in peptidoglycan cross-linking.
dTDP-4-dehydrorhamnose 3,5-epimerase (RmlC) is involved in the biosynthesis of the dTDP-L-rhamnose which is required for connection of the galactan region of arabinogalactan to peptidoglycan molecule.
Enoyl-[acyl-carrier-protein] reductase [NADH] (InhA) is involved in the biosynthesis of mycolic acids playing a key role in the fatty acid elongation process.
Decaprenylphosphoryl-beta-D-ribose oxidase (Dpre1) is a component of the complex that catalyzes the formation of decaprenyl-phospho-arabinose (DPA), a key precursor required for the synthesis of cell-wall arabinans.
N-Acetylglucosamine-1-phosphate uridyltransferase (GLMU) is an enzyme that catalyzes the final two steps in the biosynthesis of UDP-GlcNAc and is essential for the synthesis of the lipid A of lipopolysaccharide (LPS).
Shikimate kinase (AroK) is necessary for the synthesis of the common precursor of aromatic amino acids and secondary metabolites by phosphorylation of the 3-hydroxyl group of shikimic acid using ATP. The amino acids biosynthesis is also a very attractive direction for drug design, particularly as it is represented with a broad family of enzymes.
Beta-lactamase responsible for resistance to beta-lactam antibiotics.
Isocitrate lyase is in charge of the first step of the glyoxylate shunt, namely the synthesis of C4 dicarboxylic acids from C2 compounds.
ATPases (ClpC, ClpX) – chaperon proteins that induce degradation of proteins by bacterial proteasome. Play an essential role in bacterial proteostasis.
Mtb regulatory Serine Proteases (ClpP1P2) – serine proteases that form bacterial proteasomes and are responsible for protein homeostasis and degradation in bacteria.
Peptidyl tRNA hydrolase (Pth) is an essential Mtb-specific RNA hydrolase that participte in bacteria homeostasis and protein synthesis control.
The vHTS procedure assumes the application of grid-based representation of the binding site and designation of constraints based on literature data (mutation experiments) or ligand-protein interaction map with subsequent partial or full matching. Besides the rating of compounds by empirical docking score, the result conformations were also visually checked for the reliability of the binding mode. At the same time, the number of reference compounds against selected targets is relatively small and possesses a low degree of diversity inside each target-based reference set.
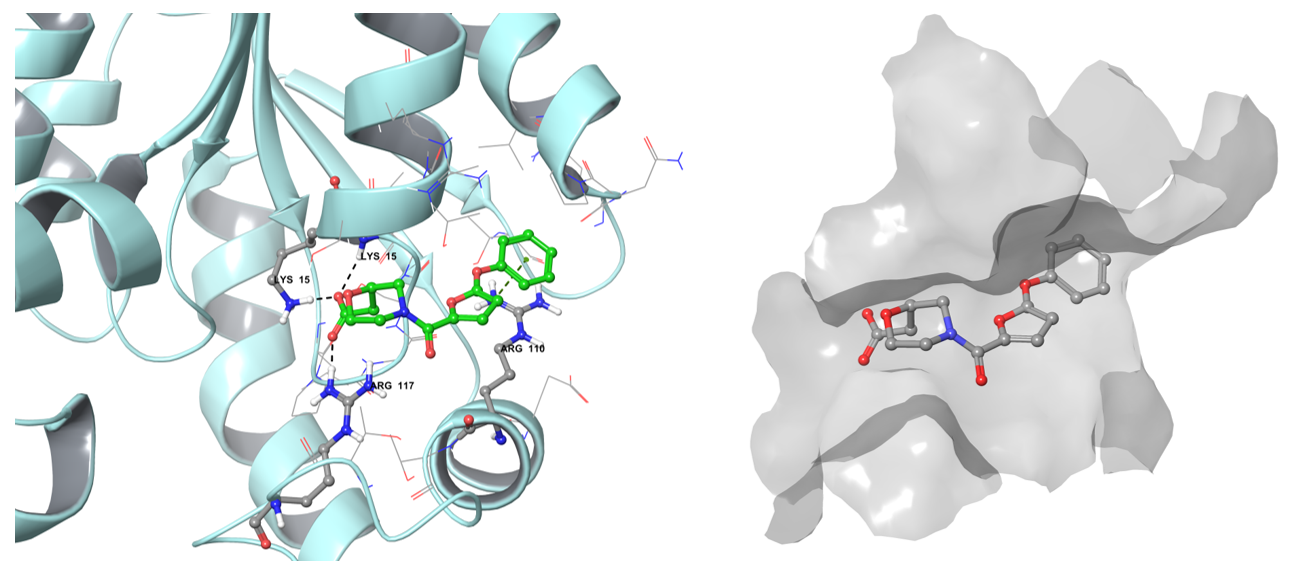
The example of the binding pose of Z1603549506 in the Shikimate kinase binding site.
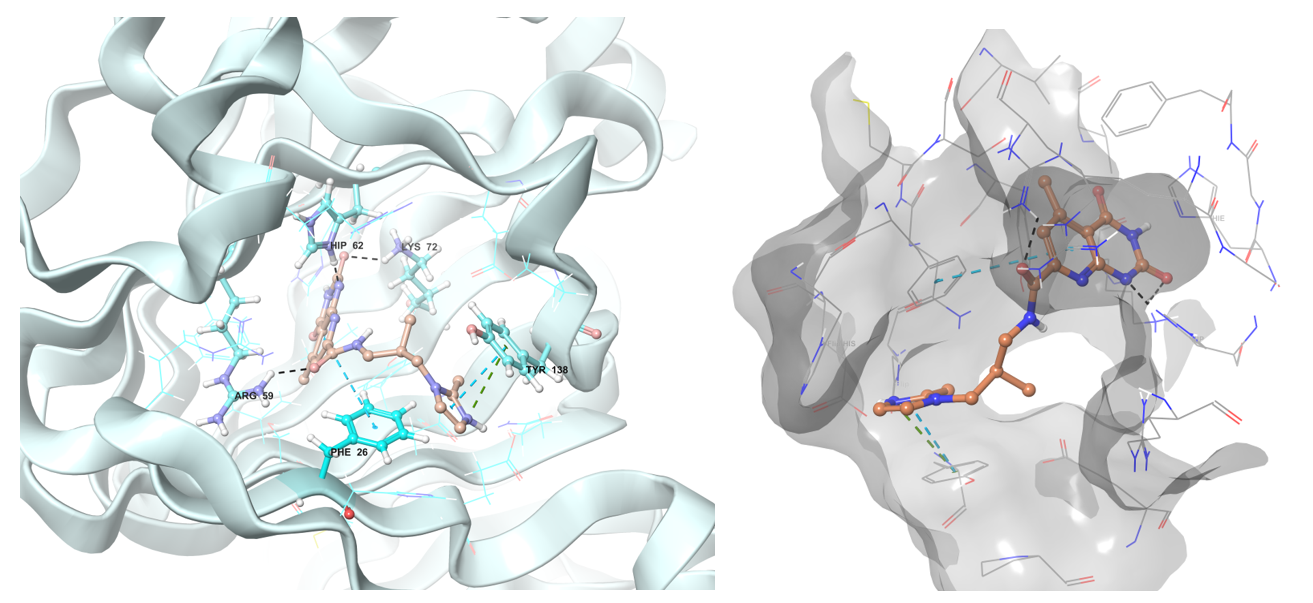
Predicted binding conformation in ribbon and shape protein representation of Z1313265676 to RmlC reductase.
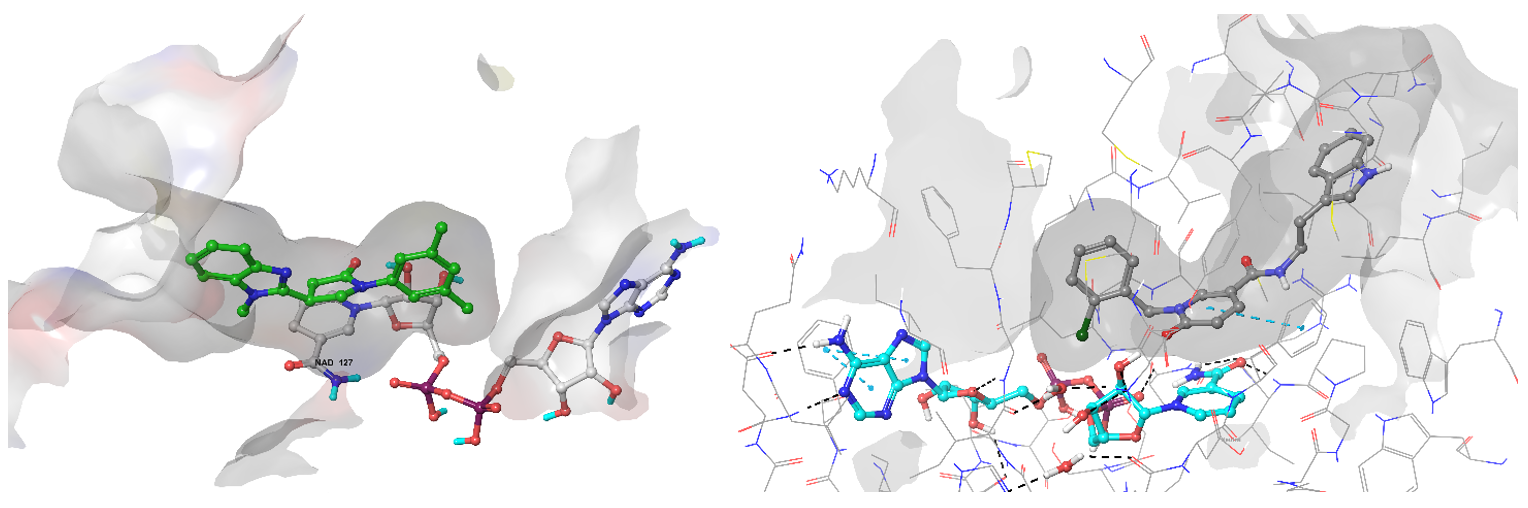
Examples of predicted hits Z111780734 (left) and Z189105794 binding conformation in the active site of InhA enzyme.
References
-
Consolidated Guidelines on Drug-resistant Tuberculosis Treatment
World Health Organization, 2019. Available online: https://www.who.int/tb/publications/2019/consolidated-guidelines-drug-resistant-TB-treatment/en/ -
In Silico Strategies in Tuberculosis Drug Discovery
Macalino, S. J. Y.; Billones, J. B.; Organo, V. G.; Carrillo, M. C. O. Molecules2020, 25(3), 665. doi:10.3390/molecules25030665 -
Drug Resistance Characteristics of Mycobacterium tuberculosis Isolates From Patients With Tuberculosis to 12 Antituberculous Drugs in China
Wu, X.; Yang, J.; Tan, G.; Liu, H.; Liu, Y.; Guo, Y.; Gao, R.; Wan, B.; Yu, F. Front. Cell Infect. Microbiol.2019, 9, 345. doi:10.3389/fcimb.2019.00345 -
Mycobacterium enoyl acyl carrier protein reductase (InhA): A key target for antitubercular drug discovery
Prasad, M. S.; Bhole, R. P.; Khedekar, P. B.; Chikhale, R. V. Bioorg. Chem.2021, 115, 105242. doi:10.1016/j.bioorg.2021.105242 -
Structural Basis for Inhibition of Enoyl-[Acyl Carrier Protein] Reductase (InhA) from Mycobacterium tuberculosis
de Ávila, M. B.; Bitencourt-Ferreira, G.; de Azevedo, W. F. Curr. Med. Chem.2020, 27(5), 745-759. doi:10.2174/0929867326666181203125229 -
Discovery of New and Potent InhA Inhibitors as Anti-tuberculosis Agents: Structure Based Virtual Screening Validated by Biological Assays and X-ray Crystallography
Kamsri, P.; Hanwarinroj, C.; Phusi, N.; Punkvang, A. J. Chem. Inf. Model.2019, doi:10.1021/acs.jcim.9b00918 -
New InhA Inhibitors Based on Expanded Triclosan and Di-Triclosan Analogues to Develop a New Treatment for Tuberculosis
Chetty, S.; Armstrong, T.; Sharma Kharkwal, S. Pharmaceuticals2021, 14(4), 361. doi:10.3390/ph14040361 -
Drug discovery in tuberculosis. New drug targets and antimycobacterial agents
Campaniço, A.; Moreira, R.; Lopes, F. Eur. J. Med. Chem.2018, 150, 525-545. doi:10.1016/j.ejmech.2018.03.020 -
Recent Progress in the Development of Novel Mycobacterium Cell Wall Inhibitor to Combat Drug-Resistant Tuberculosis
Belete, T. M. Microbiol. Insights2022. doi:10.1177/11786361221099878 -
Virtual screening for identifying a putative inhibitor of rmlc, a major target protein in tuberculosis disease
Sunilkumar, B.; Basheera, S. Int. J. Pharma Bio Sci.2015, 6, 616-628. -
Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
Cross, A.; Roy, S. J. Biol. Chem.2022, 298(5), 101903. doi:10.1016/j.jbc.2022.101903 -
High-resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme
Dong, C.; Major, L. L.; Allen, A.; Blankenfeldt, W. Structure2003, 11(6), 715-723. doi:10.1016/s0969-2126(03)00098-4 -
Synthetic molecules as DprE1 inhibitors: A patent review
Imran, M.; A S, A.; Thabet, H. K.; Abida; Bakht, M. A. Expert Opin. Ther. Pat.2021, 31(8), 759-772. doi:10.1080/13543776.2021.1902990 -
Inhibiting Mycobacterium tuberculosis within and without
Cole, S. T. Philos. Trans. R. Soc. B2016, 371, 20150506. doi:10.1098/rstb.2015.0506 -
Advances in Key Drug Target Identification and New Drug Development for Tuberculosis
Mi, J.; Gong, W.; Wu, X. Biomed. Res. Int.2022, Article ID 5099312. doi:10.1155/2022/5099312 -
Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages
Rengarajan, J.; Bloom, B. R.; Rubin, E. J. Proc. Natl. Acad. Sci. U. S. A.2005, 102(23), 8327-8332. doi:10.1073/pnas.0503272102 -
Identification of cell wall synthesis inhibitors active against Mycobacterium tuberculosis by competitive activity-based protein profiling
Li, M.; Patel, H. V.; Cognetta, A. B. 3rd; et al. Cell Chem. Biol.2022, 29(5), 883-896.e5. doi:10.1016/j.chembiol.2021.09.002 -
Characterization of M. tuberculosis SerB2, an essential HAD-family phosphatase, reveals novel properties
Yadav, G. P.; Shree, S.; Maurya, R.; et al. PLoS One2014, 9(12), e115409. doi:10.1371/journal.pone.0115409 -
High throughput screen identifies small molecule inhibitors specific for Mycobacterium tuberculosis phosphoserine phosphatase
Arora, G.; Tiwari, P.; Mandal, R. S.; et al. J. Biol. Chem.2014, 289(36), 25149-25165. doi:10.1074/jbc.M114.597682 -
Regulatory Mechanism of Mycobacterium tuberculosis Phosphoserine Phosphatase SerB2
Grant, G. A. Biochemistry2017, 56(49), 6481-6490. doi:10.1021/acs.biochem.7b01082 -
Targeting the Serine Pathway: A Promising Approach against Tuberculosis?
Haufroid, M.; Wouters, J. Pharmaceuticals2019, 12(2), 66. doi:10.3390/ph12020066 -
Characterization of M. tuberculosis SerB2, an essential HAD-family phosphatase, reveals novel properties
Yadav, G. P.; Shree, S.; Maurya, R.; et al. PLoS One2014, 9(12), e115409. doi:10.1371/journal.pone.0115409 -
Identification of cell wall synthesis inhibitors active against Mycobacterium tuberculosis by competitive activity-based protein profiling
Li, M.; Patel, H. V.; Cognetta, A. B. 3rd; et al. Cell Chem. Biol.2022, 29(5), 883-896.e5. doi:10.1016/j.chembiol.2021.09.002
