A set of compounds focused on targeting molecular chaperones
2 468 compounds
Protein folding is very important as attractive field in new drug development paradigm. Control of these processes in the cell achieves through action of assembly of enzymes known as molecular chaperones. This set of diverse protein families assists a large variety of processes involving folding, translocation, unfolding, disaggregation and homeostasis of proteins within the cellular environment. In spite of intensive studies, the spectrum of cellular substrates and functions mediated by these different chaperones remains largely undefined. Meanwhile, targeting molecular chaperones is proven to be crucial for the prevention of the many deleterious effects of protein misfolding and aggregation, which might lead in the end in cell death, in neurodegeneration and in other protein misfolding diseases.
Enamine has been following investigation on molecular chaperones for years and developed a library of 2 468 synthetic compounds potentially targeted molecular chaperones. The emphasis has been made on most promising and studied targets: Heat shock proteins (Hsp90, Hsp82, Hsp27), Chaperone activity of bc1 complex-like and histone-chaperone ASF1a complex (ASF1-histone interaction). In addition, the library was enriched with bioisosteric replacement and compounds bearing new chemotypes predominantly with polar scaffolds and cores to increase novelty of the targeted sets.
The library is available in convenient and ready to ship pre-plated formats. Also, the library can be delivered in any other custom formats within a week only.
Typical Formats
Molecular Chaperones Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
MCL-2468-0-Z-10
Compounds
2 468
2 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
MCL-2468-10-Y-10
Compounds
2 468
8 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
MCL-2468-50-Y-10
Compounds
2 468
8 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
MCL-2468-10-Y-10 screening library 2 468 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD file
Library design
- Ligand based approach: pharmacophore searches, bioisosteric replacement, 3D shape-based screening.
- Protein/target-based: Docking, in silico screening with protein-ligand key interaction feature constraints.
- Strict MedChem filters including PAINS and other industry affiliated structural filters and rules.
The sets of the reported active molecules, collected for the targets mentioned above, were carefully analyzed. Series of 3D pharmacophore models within volume restriction constraints were created and further validated with the set of reference actives and non-active ligands (Figure 1). Enamine’s MedChem stock compound collection (Ro5 compliant and filtered through series of MedChem filters) was then screened against these models. The results were inspected visually, and molecules derived from trivial chemotypes, as well as those poorly matching the pharmacophore models, were removed. The bioisosteric replacement of the core structures and selection of compounds by privileged motifs were also used to enrich the library with new valuable structures and prospective drug/lead-like compounds.
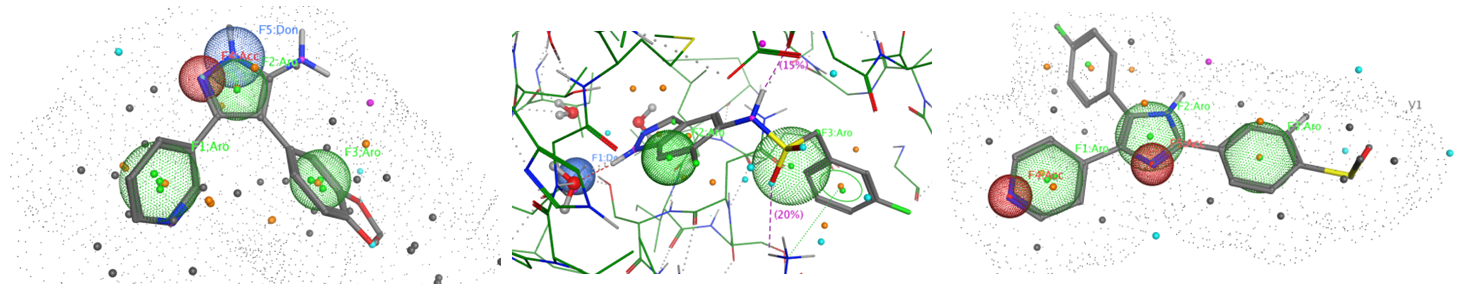
Figure 1. Examples of 3D pharmacophore models. In the cases when protein 3D structure was known superposition of ligand and protein key features was used to create pharmacophore model.