Designed for the discovery of new SARS-CoV-2 and pan-Coronavirus antivirals
16 800 compounds
In spite of the previously known coronavirus outbreaks in 2002 and 2003 followed later in 2012 with MERS-CoV spread in middle-Asia, no effective treatment has been developed. Shortage of financial support and attention in R&D area resulted in a desperate outcome. The world appeared to be largely unprepared for the new outbreak, caused by SARS-CoV-2, which began in December 2019 and quickly grew into a global pandemic. The past research on SARS/MERS-CoV family of viruses has not produce any lead series which can be quickly progressed into marketed drugs.
An unprecedented biotech race is now on to design cures for COVID-19 and to curb the crisis until a reliable vaccine is available and widely adopted. While most of the efforts are focused on drug repurposing strategies, de-novo drug discovery is essential for achieving sufficient specificity and efficacy towards the new SARS-CoV-2 virus.
To meet the urgent need for potent new antivirals against COVID-19, Enamine has designed and preplated for quick delivery a small-molecule compound library focusing on SARS-CoV most promising targets. This library incarnates Enamine’s rich experience in design and synthesis of antiviral compounds. We take an active part in the global open science initiative COVID Moonshot project, providing synthesis, ADME-Tox profiling, medicinal chemistry and logistic services.
The Coronavirus Library consists of 7 sublibraries which can be also acquired separately.
Typical Formats
Coronavirus Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
COV-16800-0-Z-10
Target
3CLpro, ACE2, TRPMSS11E and all below
Compounds
16 800
14 plates
Amount
up to 300 nL of 10 mM DMSO stock solutions
Plates and formats
1536-well plates, 1280 compounds per plate, 1-4 and 44-48 columns empty
Price
Catalog No.
MPR-3440-10-Y-10
Target
Main protease Mpro, non-covalent compounds
Compounds
3 440
11 plates
Amount
10 µL of 10 mM DMSO stock solutions
Plates and formats
384-well plates Greiner Cat. No. 781280, 320 compounds per plate, 1, 2 and 23, 24 columns empty
Price
Catalog No.
MPC-2640-50-X-20
Target
Main protease Mpro, covalent compounds
Compounds
2 640
33 plates
Amount
50 µL of 20 mM DMSO stock solutions
Plates and formats
96-well plates, Greiner Cat. No. 650201, 80 compounds per plate, 1 and 12 columns empty
Price
Catalog No.
TMP-3200-25-Y-10
Target
Type-II and 11e transmembrane serine protease (TMPRSS2)
Compounds
3 200
10 plates
Amount
25 µL of 10 mM DMSO stock solutions
Plates and formats
384-well plates Greiner Cat. No. 781280, 320 compounds per plate, 1, 2 and 23, 24 columns empty
Price
Catalog No.
PLP-3200-50-Y-10
Target
Papain-like protease (PLpro)
Compounds
3 200
10 plates
Amount
50 µL of 10 mM DMSO stock solutions
Plates and formats
384-well plates Greiner Cat. No. 781270, 320 compounds per plate, 1, 2 and 23, 24 columns empty
Price
Catalog No.
RDR-2880-50-X-10
Catalog No.
RNA-dependent RNA polymerase (RdRP)
Compounds
2 880
36 plates
Amount
50 µL of 10 mM DMSO stock solutions
Plates and formats
96-well plates, Greiner Cat. No. 650201, 80 compounds per plate, 1 and 12 columns empty
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library design
To design our library we carefully collected all available structural information of most promising SARS-CoV protein targets. The protein structures were scrutinized and docking models were created. The docking models were validated by short MD simulations and verified then by ability to form complexes with reported active molecules. We have focused on the following 5 targets and NSP, reported to be most important for virus transmission and essential for viral replication:
- SARS-CoV-2 main protease Mpro (also called 3CLpro)
- RNA-dependent RNA polymerase (RdRp)
- Papain-like protease (PLpro)
- Angiotensin-converting enzyme 2 (ACE2) receptor
- Type-II transmembrane serine protease (TMPRSS2)
- Non-structual proteins (NSP) of SARS-CoV-2, with reported 3D-struture.
For cysteine and serine proteases (Mpro and TMPRSS2) covalent docking has been carried out to identify promising covalent binders, which can elongate inhibition action. The databases of screening compounds were preliminary filtered to contain only compounds with specific warheads that are selective to each amino acid:
sublibrary1
2 640 compounds (acrylamides, chloroacetamides, vinyl sulfones and beta-lactams)
sublibrary2
560 compounds (sulfonyl fluorides, N-nitriles, chloroacetamides, epoxides, boronics)
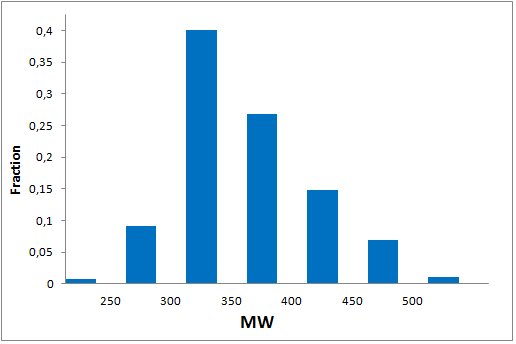
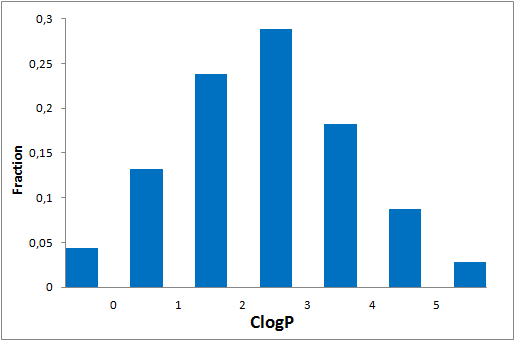
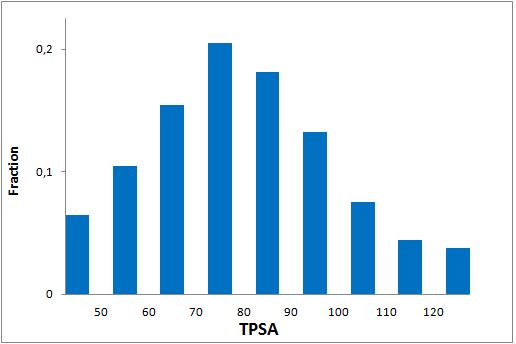
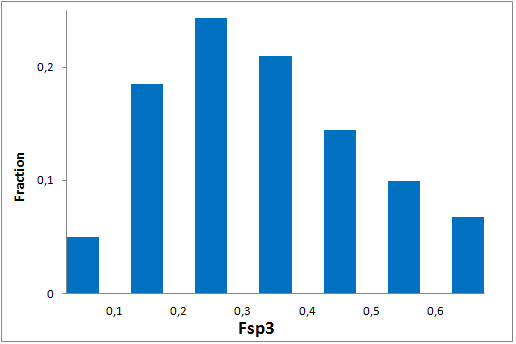
All other target-focused sublibraries passed stringent MedChem filters, including PAINS and Molecular Parameters restrictions to represent mainly lead-like space. The hits derived from our library can be easily followed with analogs and growing strategy to maximize MedChem variability within the structure.
Examples of the docking results
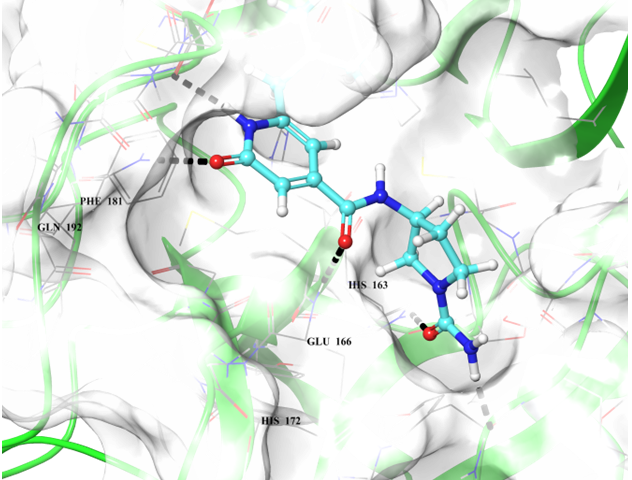
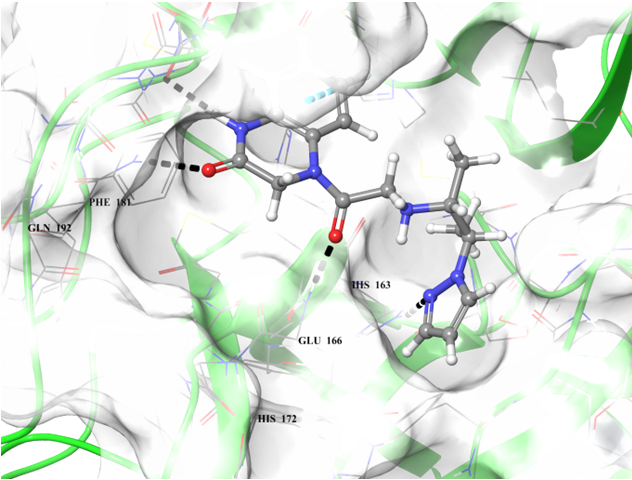
Binding poses of two predicted hit molecules after docking calculation on Mpro protease Z1609752806 (light blue ligand on the left side) and Z1143050660 (grey ligand on the right side).
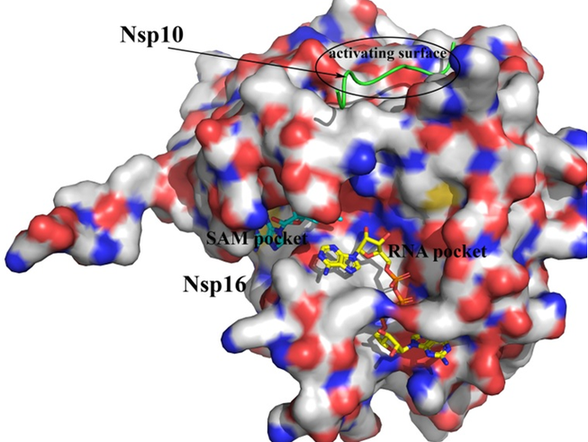
SARS-CoV-2 NSP16 protein bound to NSP10. All 3 binding pockets were used for molecular docking calculations.
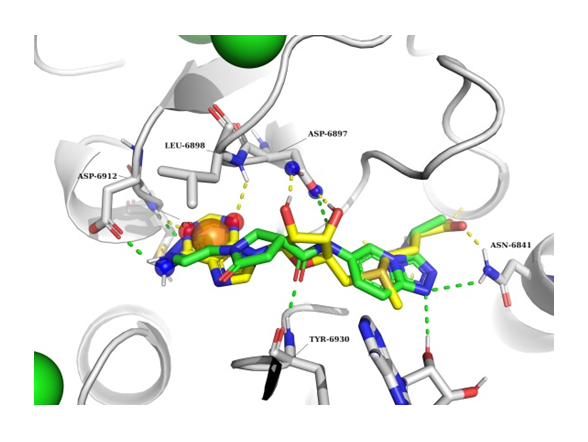
The example of NSP16 SAM binding pocket docking result. NSP16 in grey, docked ligand in green, SAM molecule in yellow.