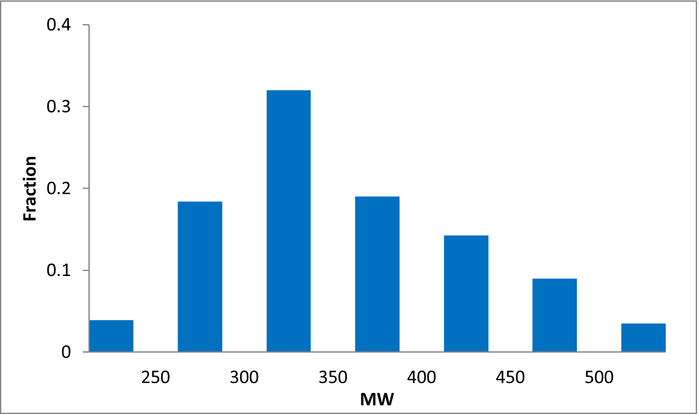
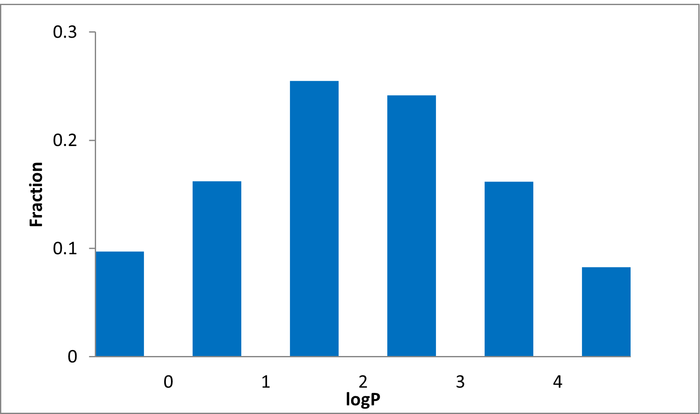
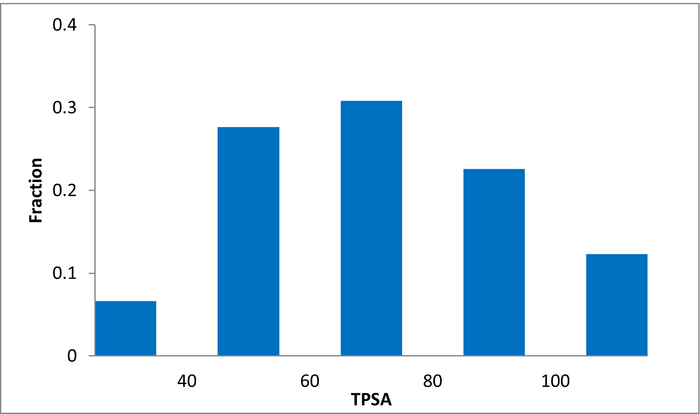
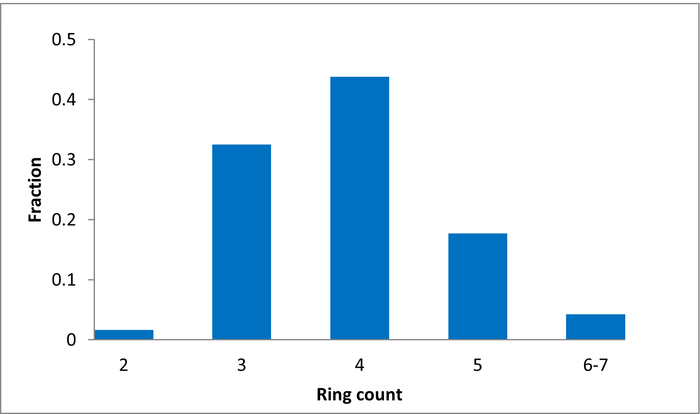
Designed for identification of new actives against proteins essential for DNA stability
5 760 compounds
For a long time, targeting DNA was considered only in the context of non-specific and cytotoxic agents. Almost all known to date DNA-interacting drugs pose catastrophic actions to the live cells. Several well-known, previously, and still widely used cancer drugs such as Cisplatin, Doxorubicin, and ethidium bromide are non-selective DNA binders or intercalating compounds. Within the last several years protein-DNA interaction interfaces gained much attention among drug hunters. This area is under investigation and requires intensive research to bring new molecules to the clinic. To meet the growing need for new specific DNA binders, we have created a versatile library combining different approaches and tools.
The library has been plated for most convenient and quick access. Using our DNA focused library you receive multiple benefits, allowing you to save on time and costs in hit expansion and optimization:
- Analogs and hit samples resupply from dry stock of over 4.7M compounds
- Straightforward & low-cost synthesis of follow-up libraries through our REAL Database technology
- Medicinal chemistry support enhanced with on-site broad ADME/T panel
Typical Formats
DNA Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
DNA-5760-0-Z-10
Compounds
5 760
5 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
DNA-5760-10-Y-10
Compounds
5 760
18 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
DNA-5760-50-Y-10
Compounds
5 760
18 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
DNA-5760-10-Y-10 screening library 5 760 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD file
Library design
Different DNA structures have been carefully investigated and analyzed: (1) G-quadruplex structures; G-quadruplexes are noncanonical DNA structures that consist of stacked G-tetrads formed by guanine-rich sequences. These structures are stabilized by monovalent cations such as K+ and Na+. G-quadruplexes are commonly found in the promoter regions of oncogenes like MYC and play an important role in regulating gene expression. Understanding the binding of ligands to G-quadruplexes provides insights into potential therapeutic strategies for targeting oncogenes and modulating gene expression. (2) The double helix DNA structure is a fundamental arrangement where two strands of DNA wind around each other. The integrity of DNA double-strands is crucial for the stability and functioning of cells and organisms. DNA double-strand breaks are particularly critical events that can lead to various consequences. They are associated with the development of human syndromes, neurodegenerative diseases, immunodeficiency, and cancer.
To search for potential DNA ligands all available in PDB and in PDBe liganded DNA structures were analyzed. Then the following entries were selected for in silico screening: 5W77, 6JJ0, 7KBW, 5T4W, 6IJW, 7KWK, 5Z80, 6CCW, 6SX3, and 7EL7. The two main approaches, binding types, were used for search of potential ligands:
- Intercalation: ligands insert itself between the stacked DNA bases pairs.
- Binding insight the DNA grove: ligands binds to the DNA groove without significantly disrupting of the DNA structure.
The molecular docking simulations were conducted using the following queries: pharmacophore of H-bond acceptors and H-bond donors, aromatic group, stearic and volume exclusion features. Each pharmacophore model includes both common and specific features, making the binding points specific to each DNA target structure and its corresponding native ligand. Several examples are provided below.
Examples of molecular docking simulation to histone lysine methyltransferases
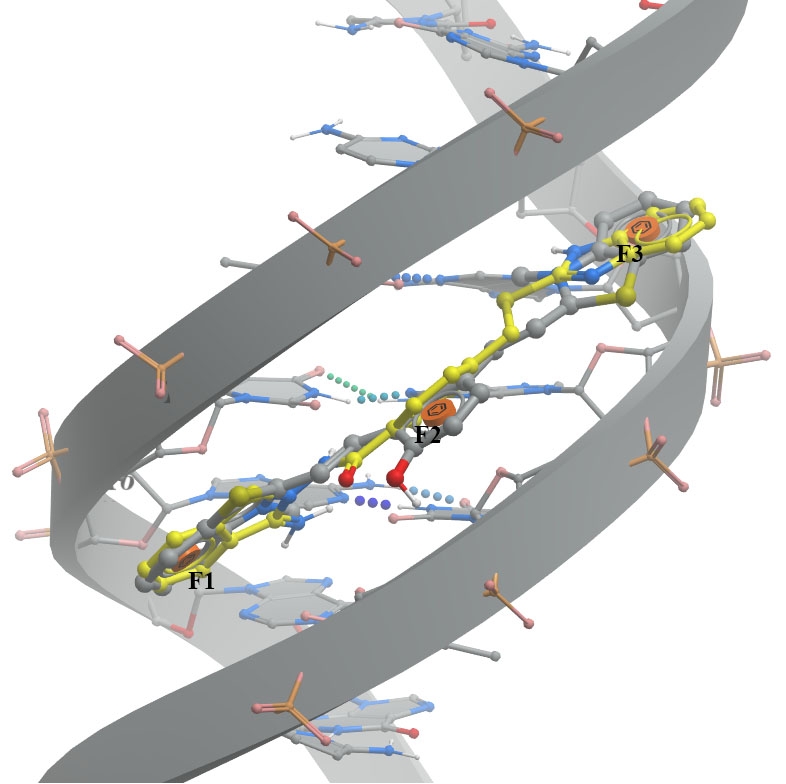
A. The minor groove (PDB ID 6IJW - left image) involves binding between QCy-DT DNA ligand and 6IJW, ligand should match with three aromatic features.
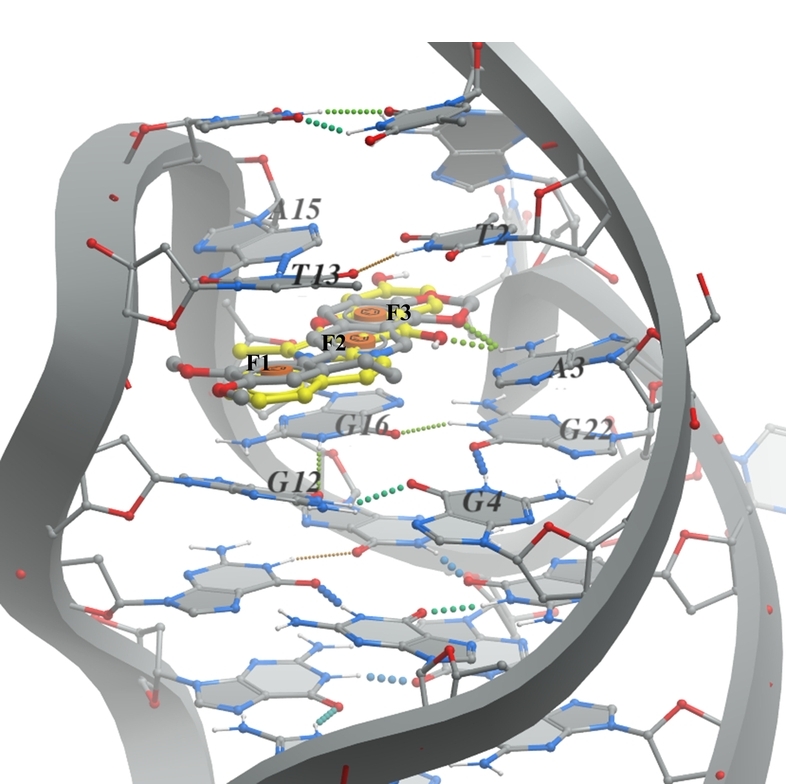
B. G-quadruplex (PDB ID 6CCW - right image) the pharmacophore model based on the presence of epiberberine intercalated within the G-quadruplex. The ligand should consist of three aromatic parts (F1, F2, and F3) that can stack between the DNA nucleobases G12, G16, A15, and T13.
DNA and its native ligand are colored in grey, docked ligands are shown in yellow.