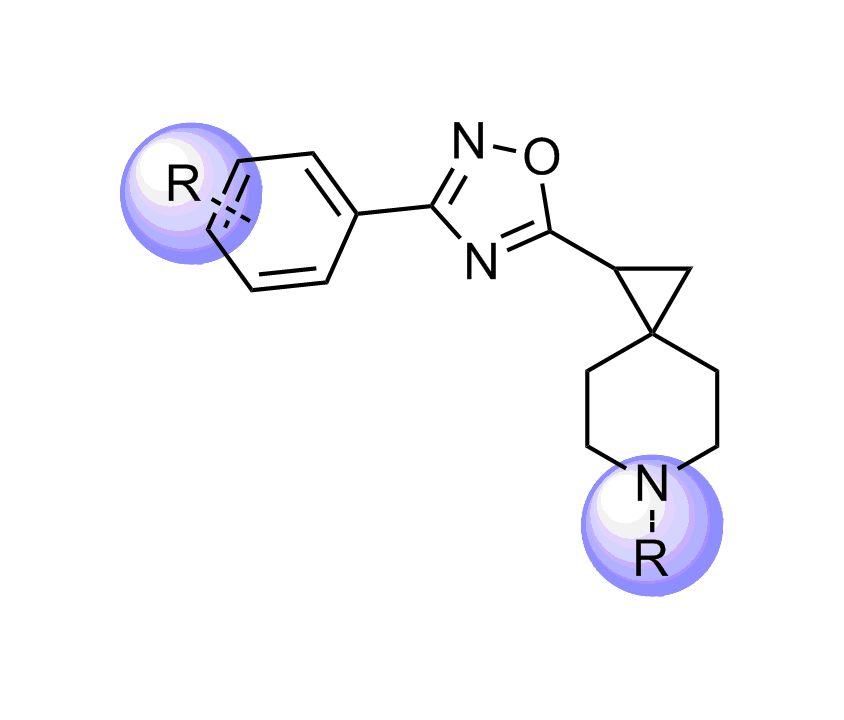
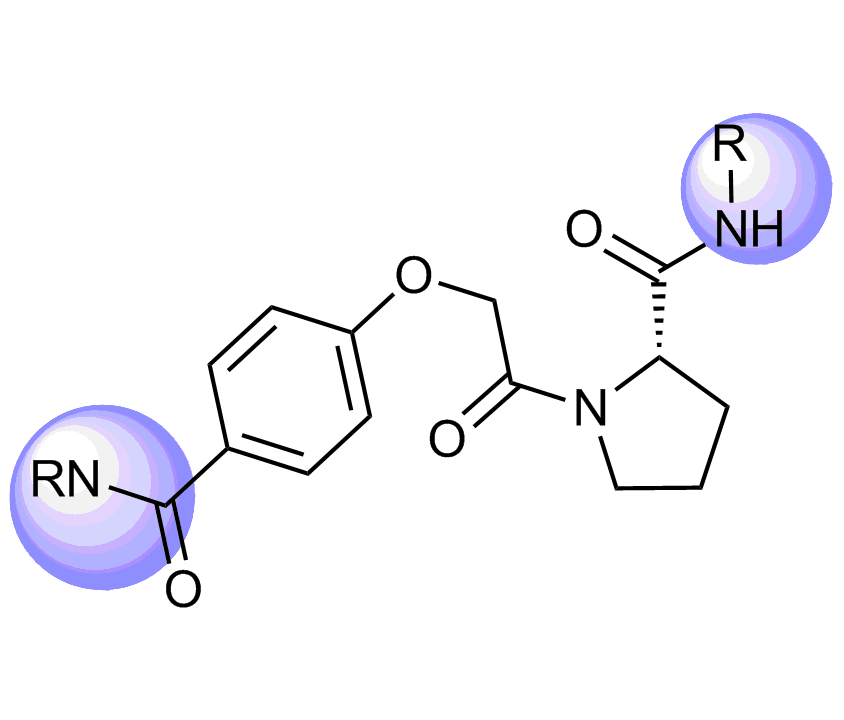
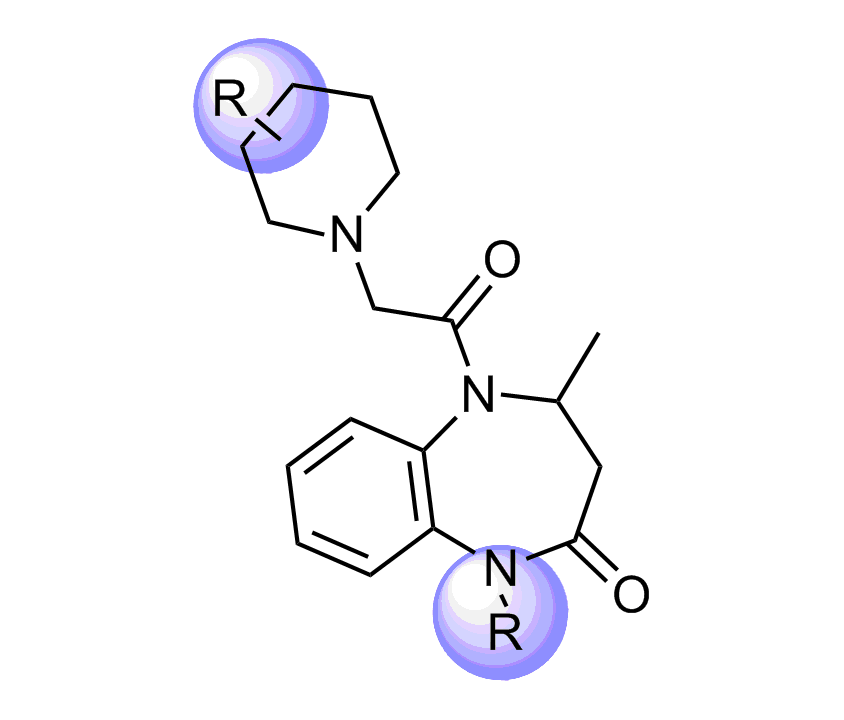
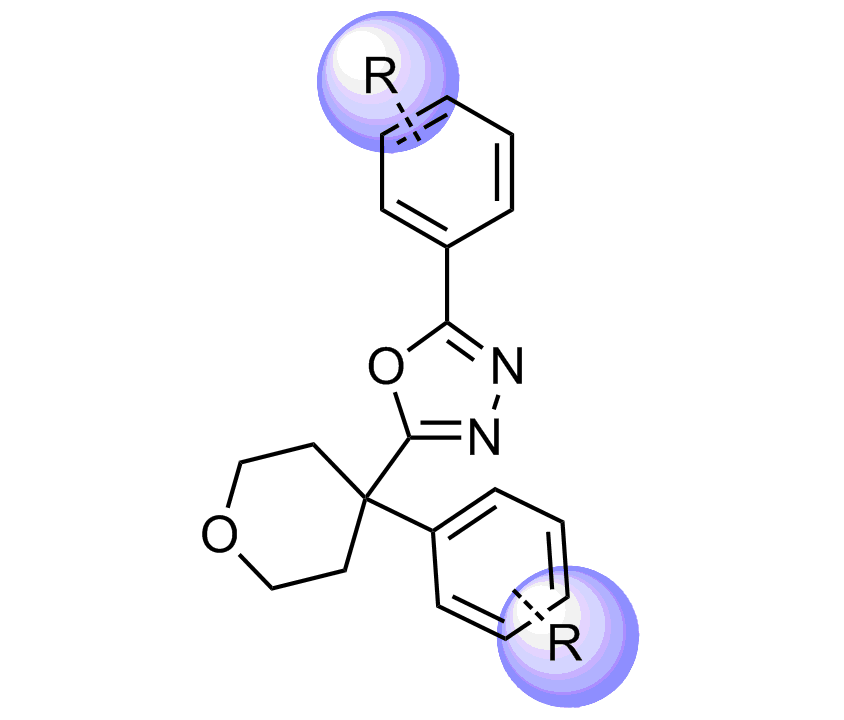
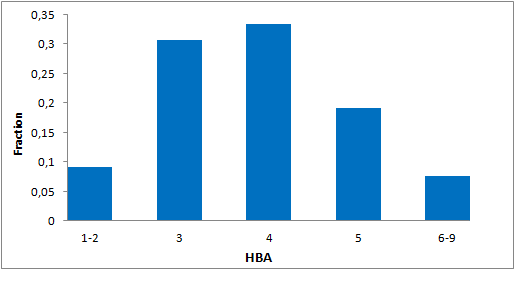
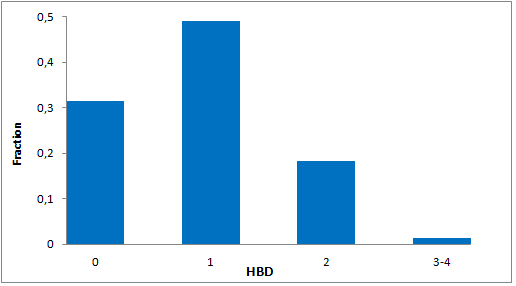
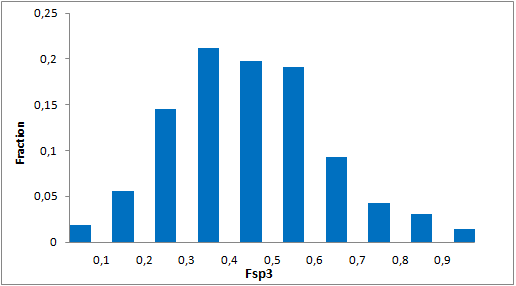
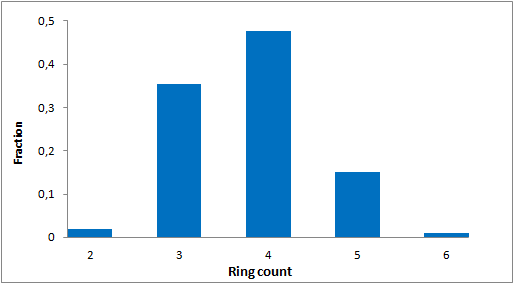
Selected molecules able to mimic common protein motifs
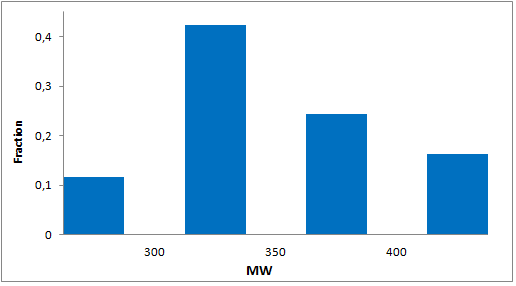
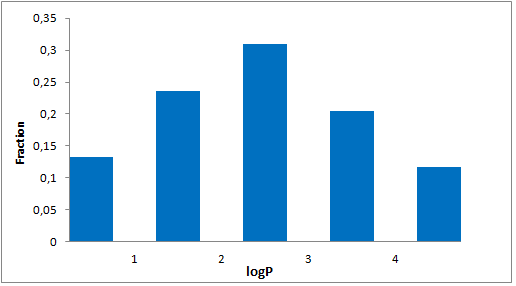
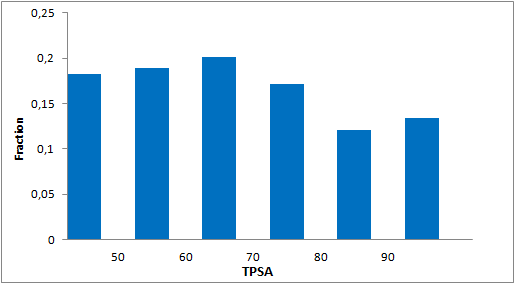
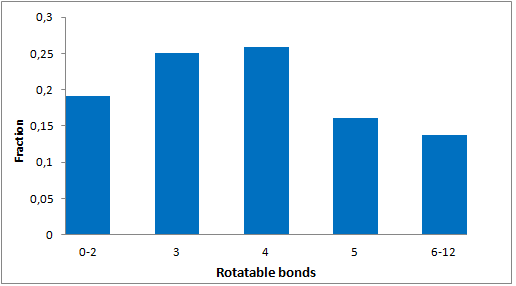
8 960 compounds
Over 250 000 individual protein-protein interactions (PPIs) were identified in human proteome. Although many of these exhibit topologically complex and formidable surface areas, in many instances the successful binding is attained via a limited number of key residues ("hot spots“). There are multiple reports in the literature describing both computational and diversity-oriented approaches to developing small-molecule modulators of PPIs. Of these, non-peptidic a-helix and b-turn imetics are of particular importance due to their key role in multiple deregulated pathways leading to cancer, neurodegenerative disorders, inflammation and immunological disorders.
Typical Formats
Protein Mimetics Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
PML-8-0-Z-10
Compounds
8 960
7 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
PML-8-10-Y-10
Compounds
8 960
28 plates
Amount
10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
PML-8-50-Y-10
Compounds
8 960
28 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
PML-8-10-Y-10 screening library 8 960 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library code: PML-8960
Version: 29 September 2023
8 960 compounds
sublibrary of PPI-40
Library design
Traditional three residue approach
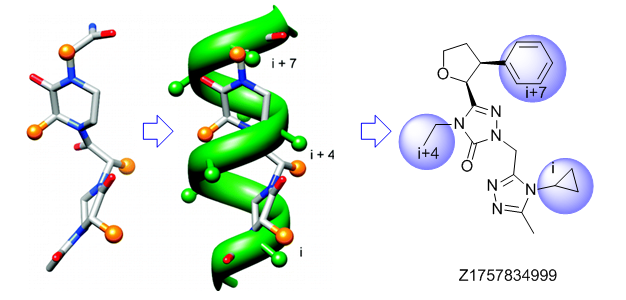
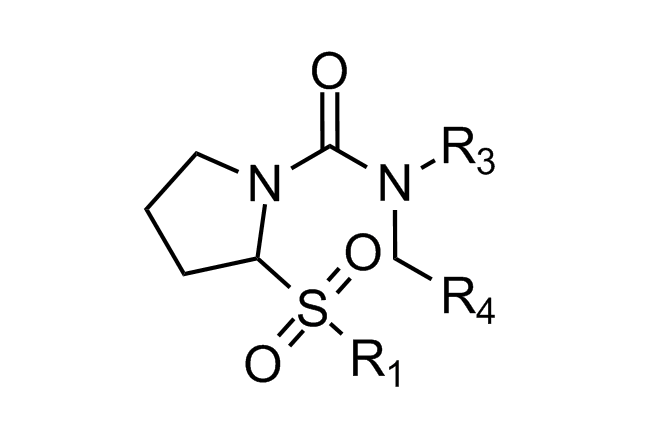
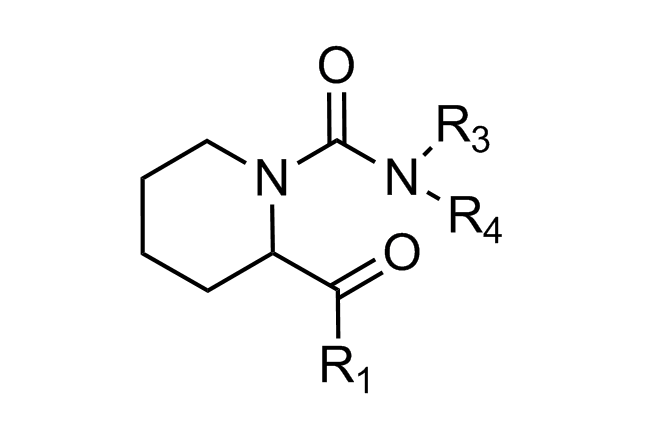
Alpha-helix Mimetics
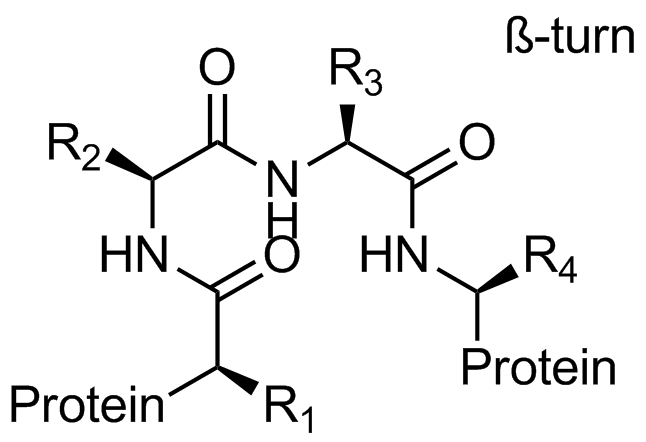
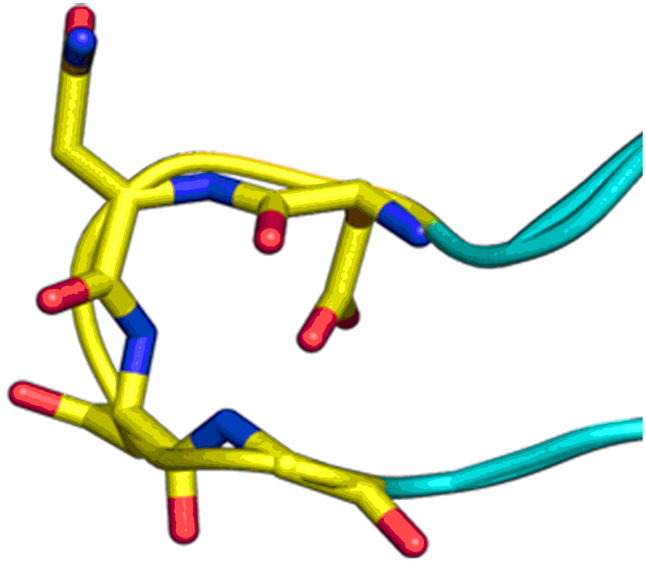
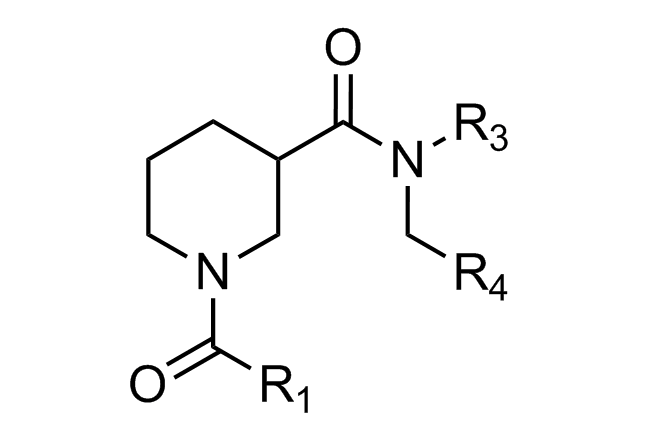
Beta-turn Mimetics
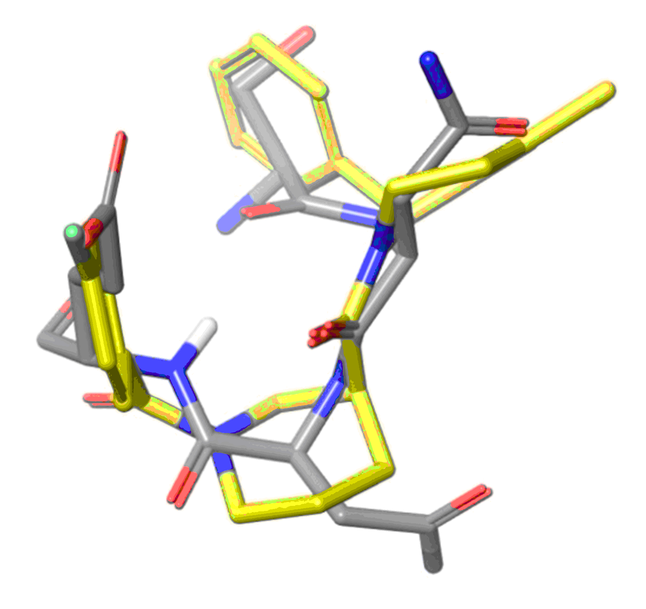
Ligand-based approach
Target based approach. We carried out design of a focused compound set targeting selected alpha-helix domains and beta-turns such as mdm2-p53, mdm2-CK1α, HIF1-α , MEF2-HDAC4, HIV-1 gp41, CD81/CV, Bcl-2/Bcl-xL, etc. After the selection by virtual screening the preference was set to the structures bearing new chemotypes.
‘Traditional’ three-residue appraoch included design of compounds with unusual cores bearing regular key-residues that mimics i, i+3/i+4 and i+7-residues located on the one recognition face (‘hot spots’). In modeling a-helix interfaces, we accounted also two-face helix mimetics. For beta-turn mimetics we applied three query models with distinct types of interaction: π-π stacking, cation-π and H-bond interaction, S-π interaction. : The latter focused design exercise was exemplified by modeling the interaction between Bim BH3 domain with Mcl-1 and Bcl-2 and identifying fused polyhydrogenated aza-heterocycles with high Fsp3 character.
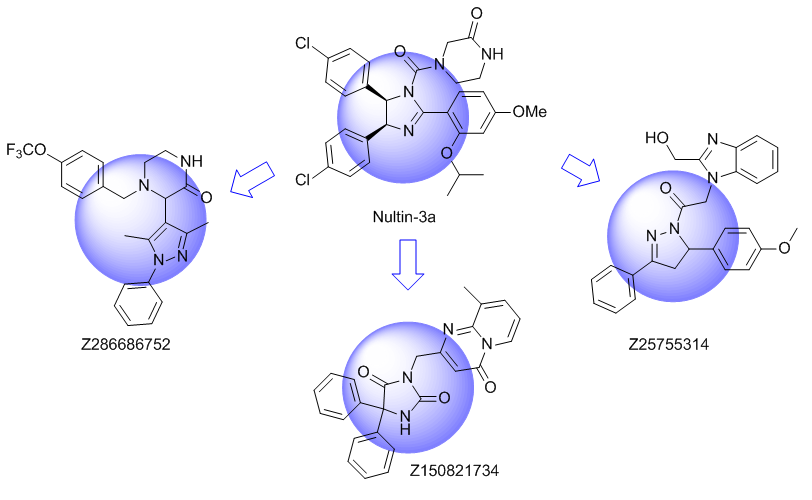
Ligand based approach. New chemotypes with close topology to known alpha-helix and beta-turns mimetics were selected. Shape based similarity and Pharmacophore screening were used as a main tools for the library design.
Unique Fsp3-enriched scaffolds exhibiting ladder-like cyclic skeletons were specially selected to enhance topological and pharmacophore interaction with the target alpha-helix and beta-turn motifs.
Examples of Selected Scaffolds in the Library