Specially selected molecules to target bromodomains
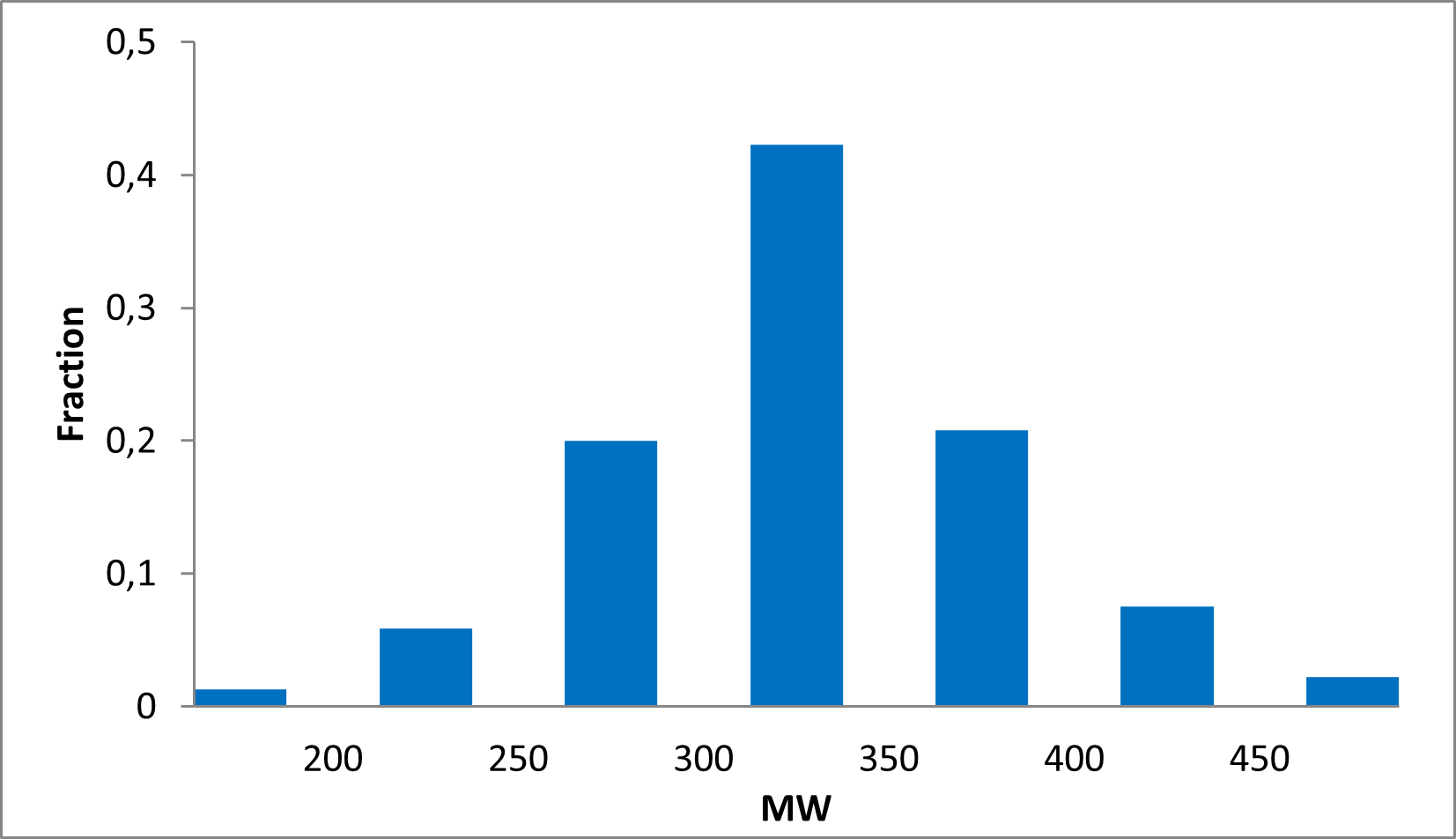
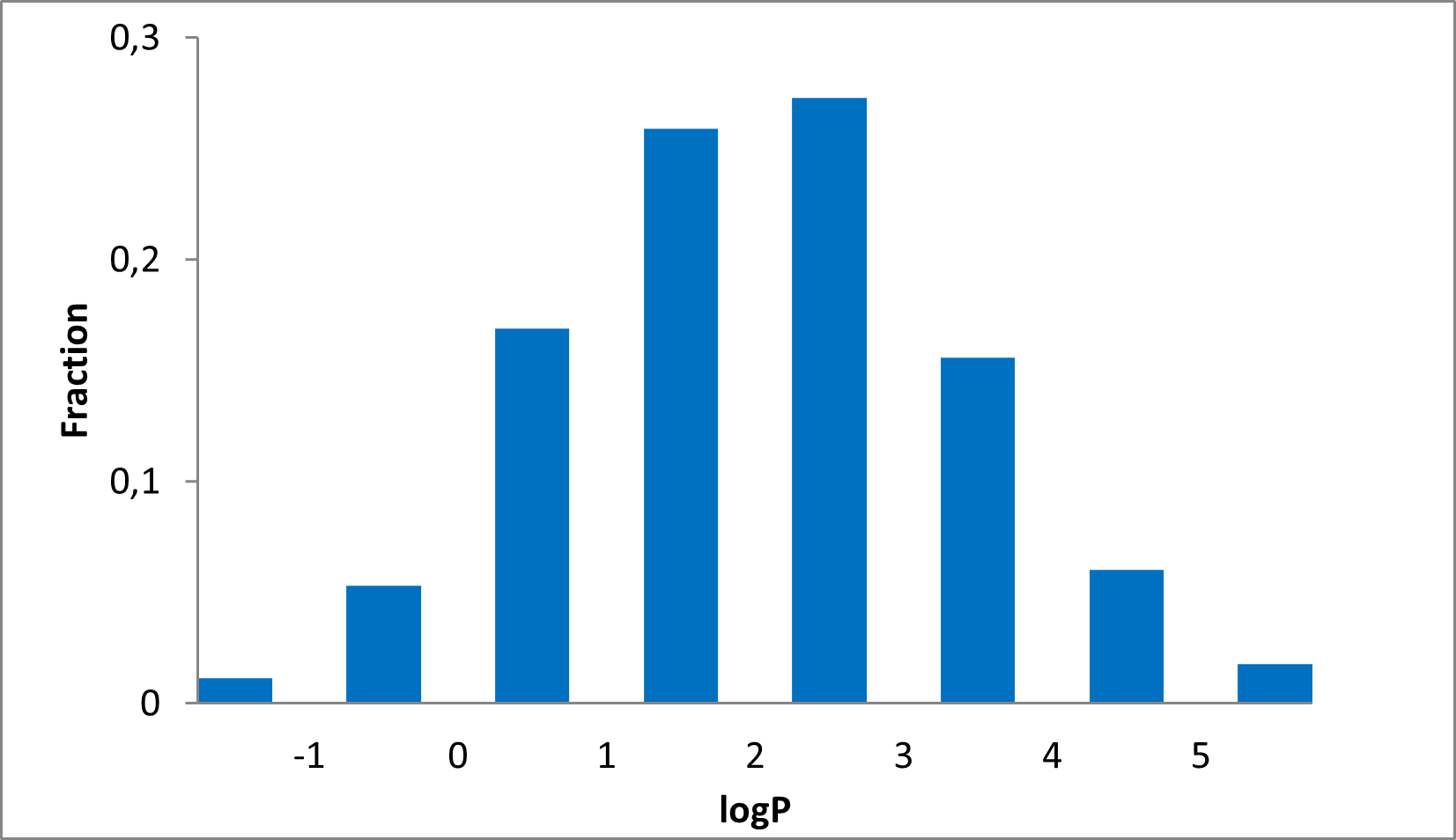
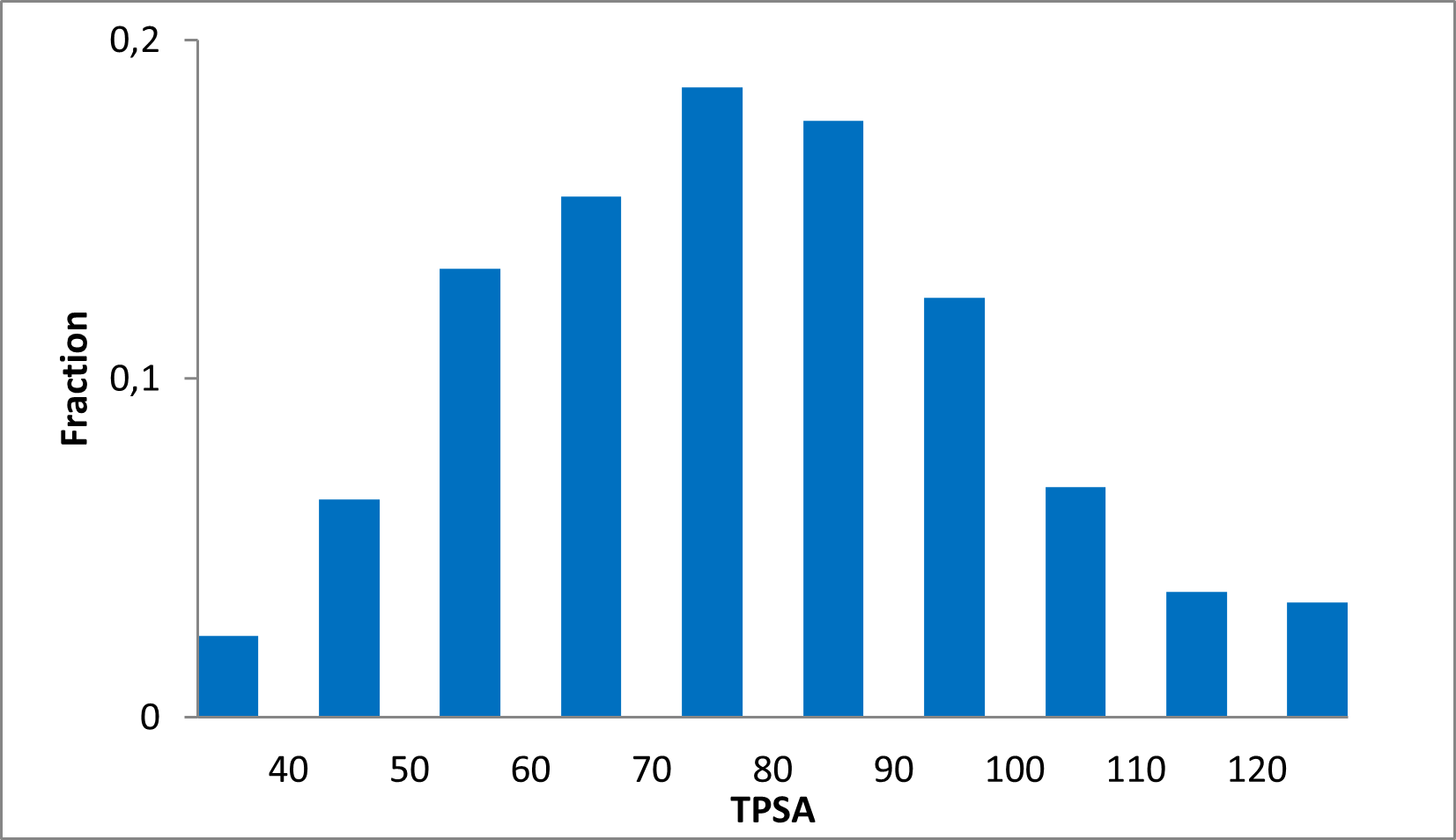
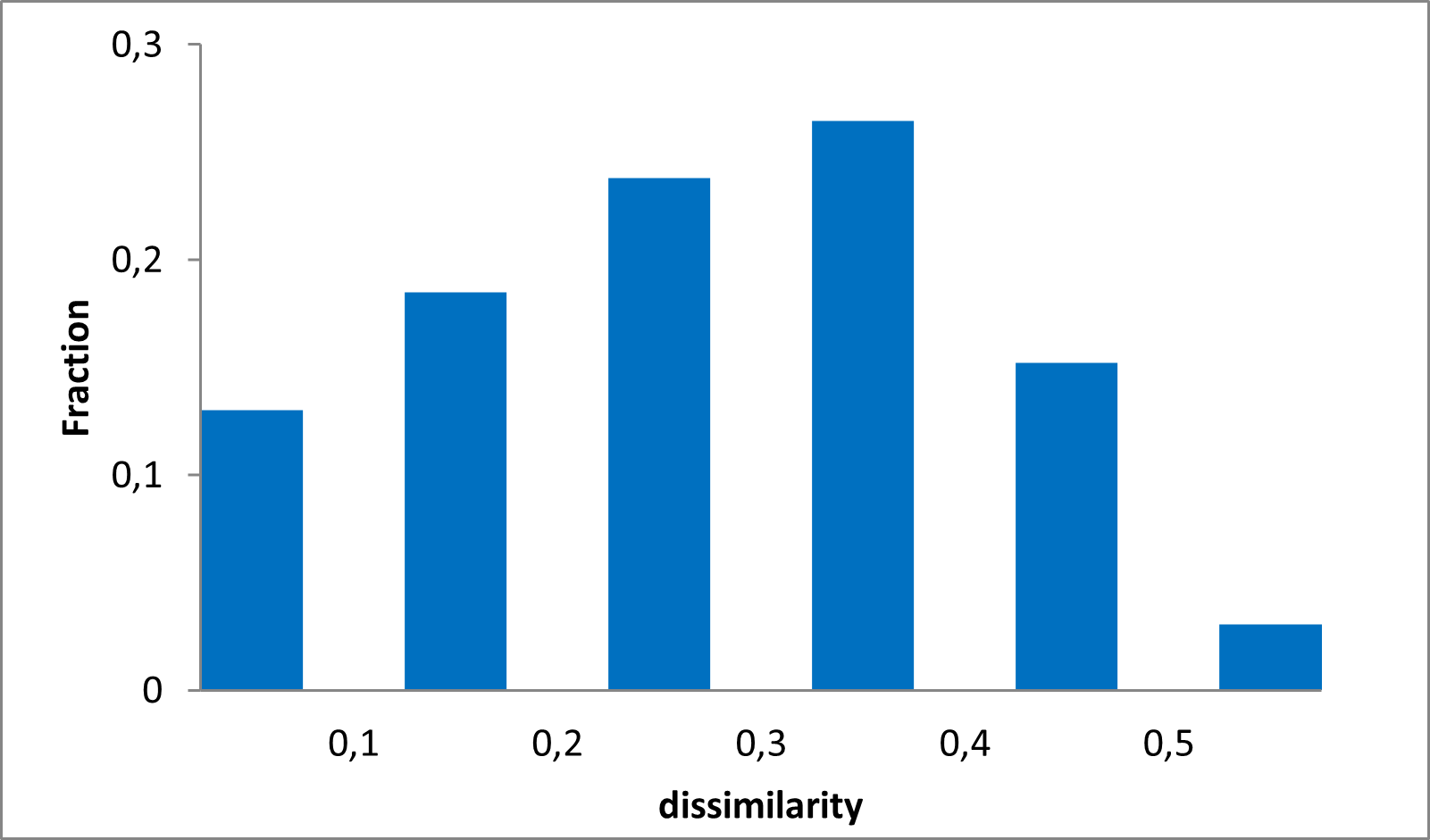
15 360 compounds
Bromodomains are protein domains found in various proteins and are involved in the recognition of acetylated lysine residues on histone proteins. These domains are named after their ability to recognize the acetyl-lysine side chain, which has a similar shape to a bromide ion. Bromodomains are essential in regulating gene expression and chromatin structure, as acetylation of histones is associated with open chromatin and active gene transcription. Dysregulation of bromodomain-containing proteins has been implicated in several diseases, including cancer and inflammation. As a result, bromodomains have become a popular target for drug discovery efforts, with several small molecule inhibitors in development for cancer and other diseases.
We focused on the search of active compounds against the most important Bromodomain families: BET subfamily: includes BRD2, BRD3, BRD4, and BRDT bromodomains, which are characterized by an extended ZA loop that interacts with acetylated lysine residues on histones. GCN5-related subfamily: This subfamily includes the GCN5 and PCAF bromodomains found in histone acetyltransferases and have a shorter ZA loop than the BET subfamily. TAF1-like subfamily: TAF1 and TAF1L bromodomains are found in transcription factor TFIID and have an N-terminal extension that interacts with other complex subunits. ATAD2-like subfamily: ATAD2 and ATAD2B bromodomains are found in AAA+ ATPases and have a long ZA loop and a unique insertion between the second and third alpha-helices. Additionally, we run docking calculations for BPTF protein that also contains bromodomain.
Typical Formats
Bromodomain Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
BRD-15-0-Z-10
Compounds
15 360
12 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
BRD-15-10-Y-10
Compounds
15 360
48 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
BRD-15-50-Y-10
Compounds
15 360
48 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
BRD-15-10-Y-10 screening library 15 360 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library code: BRD-15
Version: 7 December 2023
15 360 compounds
sublibrary of EPG-38080
Library design
We used a structure-based approach, molecular docking calculations, as the main method for the library design. All available PDB structures for each of the bromodomain subfamilies were analyzed and extracted from PDB and PDBe. Unique "protein-ligand" complexes were selected for analysis. The following PDB structures were included in our study: BRD2 (4j1p, 4a9o, 5uew, 6ffe, 7l6d, 7l9j, and 7oe8), BRD3 (3s91 and 7l9l), BRD4 (7ajn, 7axr, and 7c2z), and BRDT (4flp, 7l9a, and 7mrg); PCAF (5fdz and 5fe5); TAF1 (5i1q, 6p38, and 7jjg); ATAD2 (6veo); and BPTF (5r4i, 6lu5, and 7lp0).
All simulations were performed with a common feature: the binding of a potential ligand to Asn inside the bromodomain binding pocket. All other binding points were dependent on the specific bromodomain and the structure of its native ligand. For example, in the left picture, the docking example of 7l6d is presented. The model is characterized by the following key features: (1) an h-bond acceptor to interact with Asn 429, an HOH molecule (which creates an h-bond bridge with Tyr 386), and an h-bond acceptor in the other part of the binding pocket to interact with the peptide backbone of Asp 377; (2) two aromatic groups to fill in the subpockets between Val 435, Leu 383, and between Pro 371, Leu 381; and (3) any atom group to fill in the subpocket among Tyr 428, Asn 429, and Leu 383. In contrast, in the right picture (5fe5), the model should contain slightly different binding properties: (1) an h-bond acceptor to interact with Asn 803, an HOH molecule (which creates an h-bond bridge with Tyr 760); (2) two aromatic groups to fill in the binding pocket and potentially create stacking interactions with Tyr 809.
Examples of molecular docking simulation to bromodomains
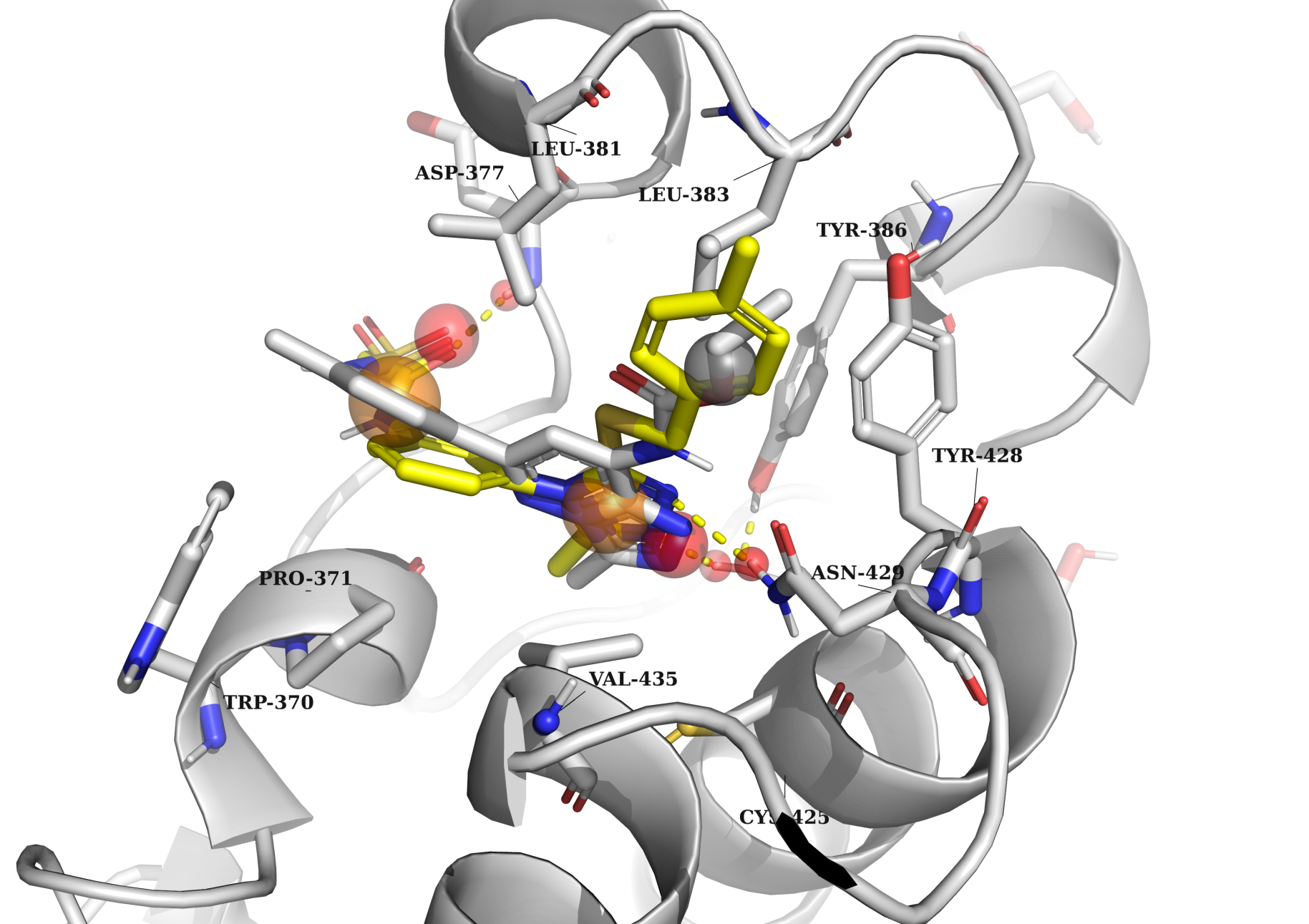
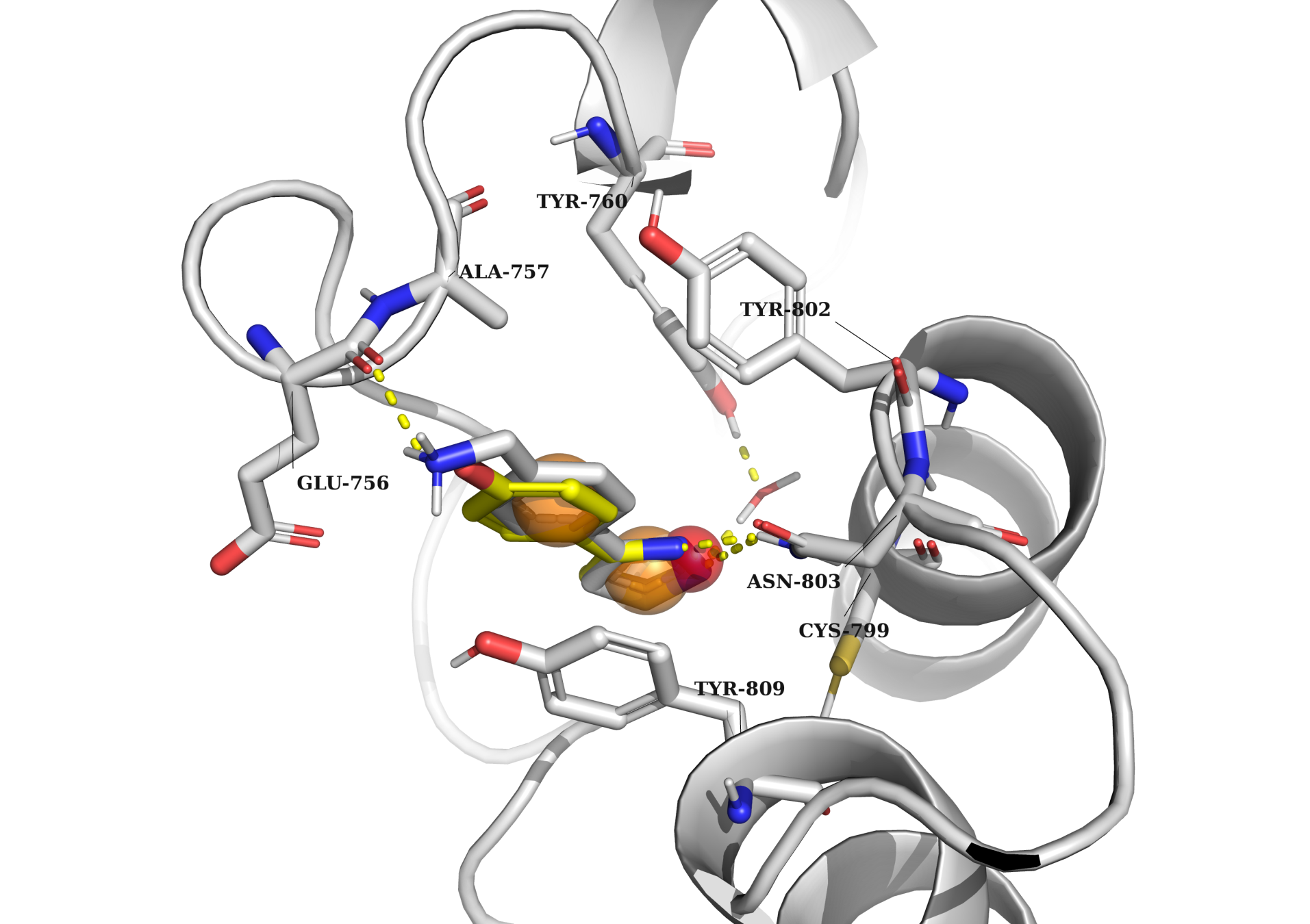
The molecular docking simulations to bromodomains were conducted using the following color scheme: red spheres represent h-bond acceptors, blue spheres represent h-bond donors, orange spheres represent aromatic groups, and grey spheres represent any other atom types. The protein and its native ligand are highlighted in grey, while an example of a docked ligand is shown in yellow.