Designed for discovery of new regulators of methabolic disorders
13 120 compounds
The kynurenine pathway is a metabolic pathway leading to the production of nicotinamide adenine dinucleotide (NAD+) from the degradation of tryptophan. Disruption in the pathway is associated with wide range of diseases and disorders including infectious diseases (e.g. HIV), neurological disorders (Alzheimer’s disease (AD), Huntington’s disease (HD) and ALS), affective disorders (schizophrenia, depression and anxiety), autoimmune diseases, peripheral conditions and malignancy. The aim of our work was to conduct searches for new potential active compounds for the kynurenine pathway, which, in turn, could be used as convenient and quality starting points for early drug development.
Typical Formats
Kynurenine Pathway Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
KYN-13-0-Z-10
Compounds
13 120
11 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
KYN-13-10-Y-10
Compounds
13 120
41 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
KYN-13-50-Y-10
Compounds
13 120
41 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
KYN-13-10-Y-10 screening library 13 120 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library code: IDO-4800
Version: 13 August 2024
4 800 compounds at 10 mM in DMSO
Library design
The search of potential actives was performed using all available protein and ligand structural data for the following targets: indoleamine dioxygenase (IDO), tryptophan dioxygenase (TDO), 3-hydroxyanthranilic acid dioxygenase (3-HAO), kynurenine aminotransferases (KATs), kynurenine 3-monooxygenase (KMO).
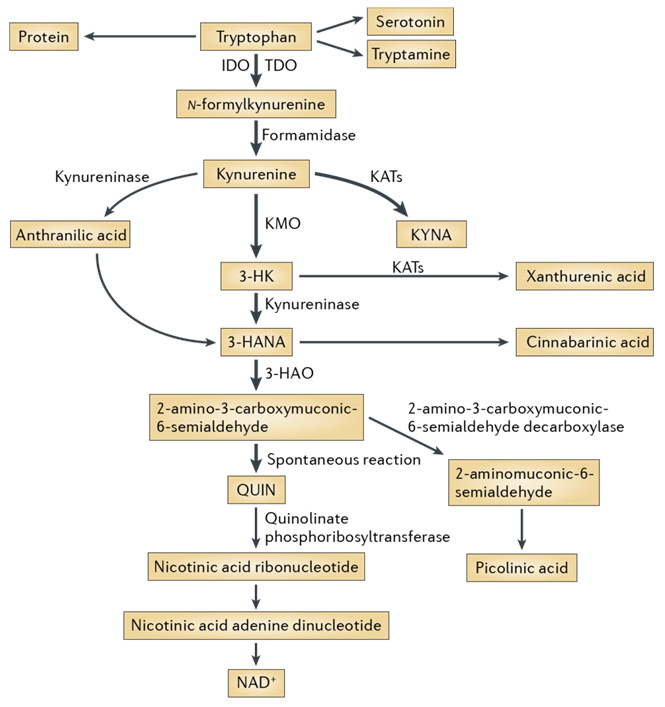
Fig. 1. The kynurenine pathway of tryptophan degradation in mammals.
Screening Models preparation and validation
All available 3D structures of the targets were retrieved from PDB. Alternate superposition and comparison of the protein structures showed high sequence identity in every separate protein target > 90 % with RMSD in a range 0.4 - 0.71 Å2. Considering all representative structures only one has been selected for model preparation taking into account ligand binding parameters (protein- native ligand) and geometric parameters of the binding sites.
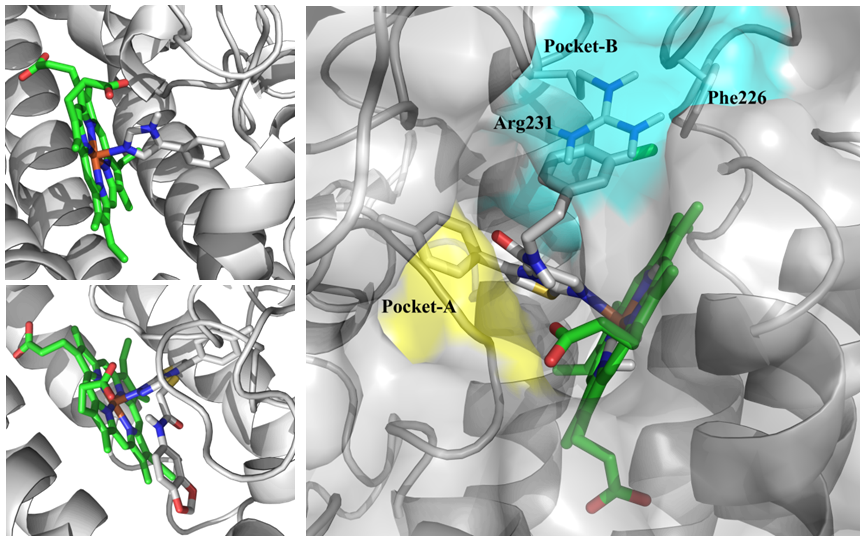
Fig. 2.Binding site of IDO1 (2D0T (top), 4PK5 (bottom) and example two main subpockets (in blue and yellow) used for molecular docking).
Molecular docking & Pharmacophore search
Basing on compounds with known activity and “protein–native ligand” complexes, ligand-based (IDO: 3, TDO: 4, KATs: 3, KMO: 2) and structure-based (IDO: 3, TDO: 6, 3-HAO: 2, KATs: 3, KMO: 4) pharmacophore models were build and validated. Drug-like Enamine database of over 1M compounds were then screened.
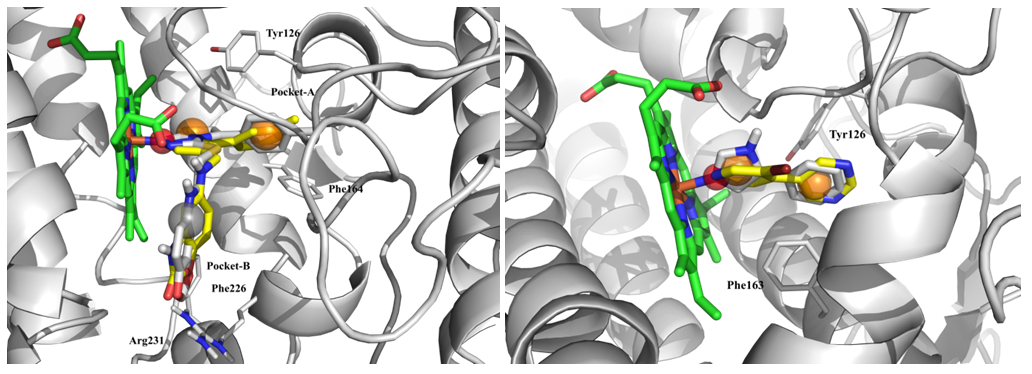
Fig. 3. Results of molecular docking: superposition of native (grey) and hit ligands (yellow) with representation of structure-based pharmacophore features.
Examples of the molecules
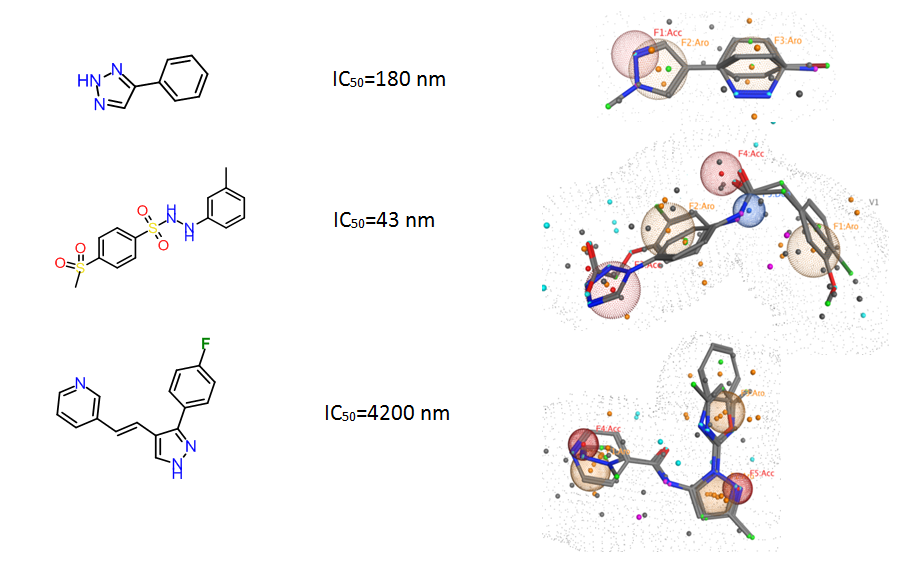
Fig. 4. Ligand-based Pharmacophore models used for in silico screening.
Number of compounds in targeted libraries after molecular docking calculation and pharmacophore searches:
Target Name
IDO
TDO
3-HAO
KATs
KMO
Number of compounds in the library
4 800
5 120
1 000
1 000
3 200
Careful analyzes of proteins and their ligand interactions involved in kynurenine pathway were performed. The iterative in silico searches using all available state-of-the-art methodologies were curried-out to yield unique sets of potentially active molecules. Chemotype control was used to enrich the libraries with structural motifs of true positives and new “patent-free” structural cores.
Developed targeted sets are intended for high-probability initial hit discovery in one step for any particular target.



