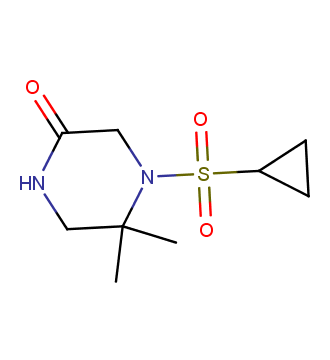
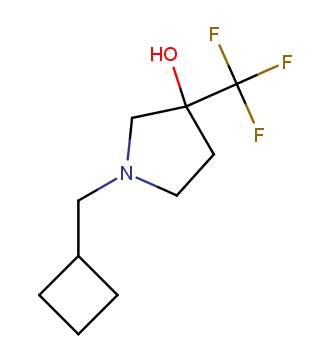
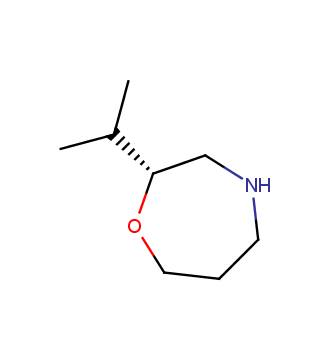
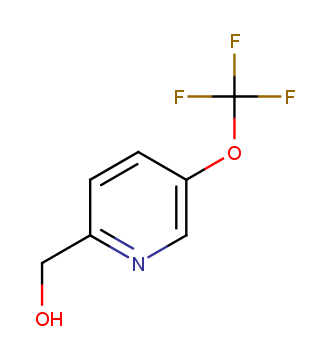
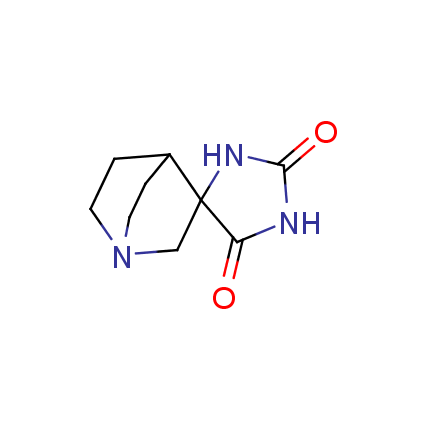
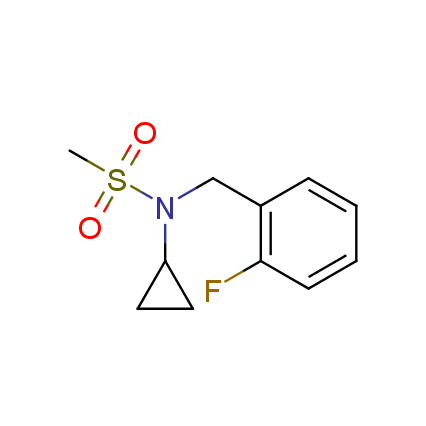
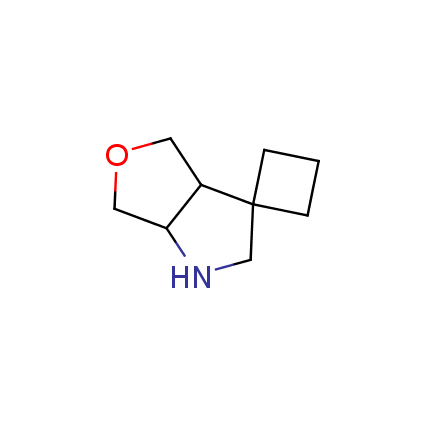
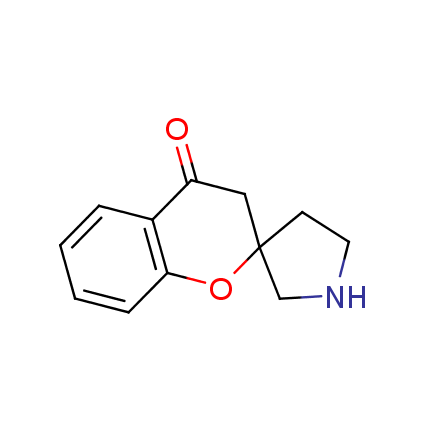
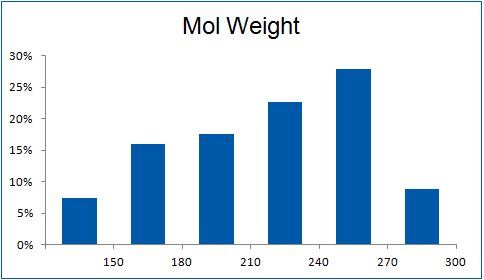
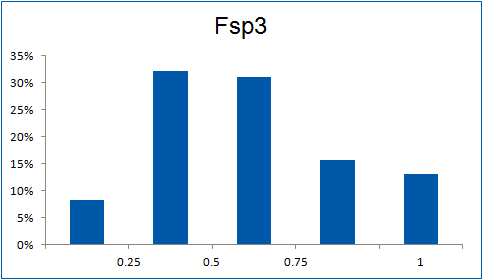
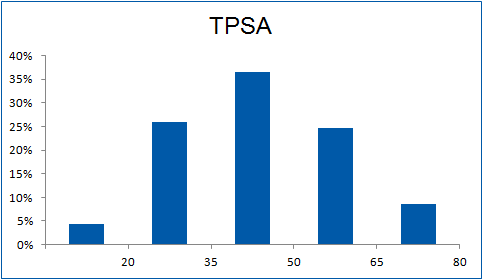
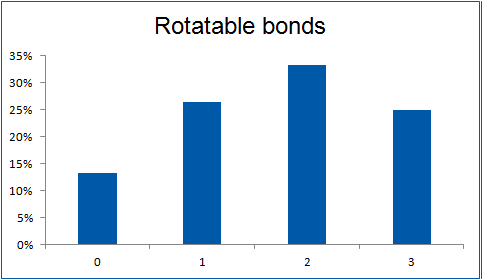
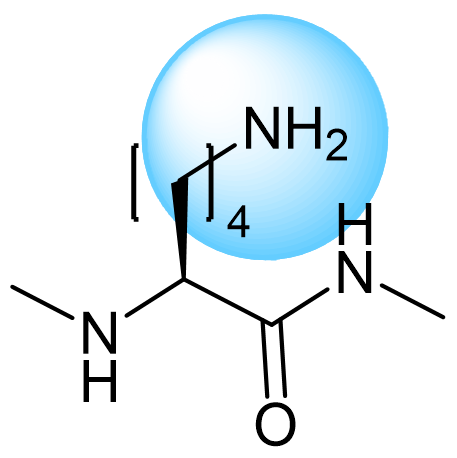
The ultimate selection of Lys-specific binders
1 600 compounds
Until recently, covalent binders targeting Lysine residue attracted less attention as compared to those modifying Cysteine or Serine. Being among the essential amino acids, Lysine can be found both at the surface and in active sites of many enzymes (e.g. kinases, viral polymerases and integrases, aldolases, DOPA decarboxylase, P-glycoprotein), and it can also participate in catalytic reactions. Moreover, Lys residue is often involved as a key player in many important signaling and metabolic processes. An example can be protein ubiquitination, which mainly occurs through lysine residues on substrate proteins or itself. Selective modification of Lys residue becomes more and more attractive for the investigation of many signaling processes in live cells and, potentially, can play a key role in the discovery of next-generation drugs.
Lysine focused Library is plated at 20 mM concentration in DMSO and is available for fast supply in any custom format. Compounds pooling based on delta MW can be provided as a formatting option. Please request the list of expected molecular weight shifts for MS-based screenings.
Typical Formats
Lysine-Focused Covalent Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
LYS-1600-0-Z-10
Compounds
1 600
2 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
LYS-1600-10-Y-10
Compounds
1 600
5 plates
Amount
10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
LYS-1600-50-Y-10
Compounds
1 600
5 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
LYS-1600-10-Y-10 screening library 1 600 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD file
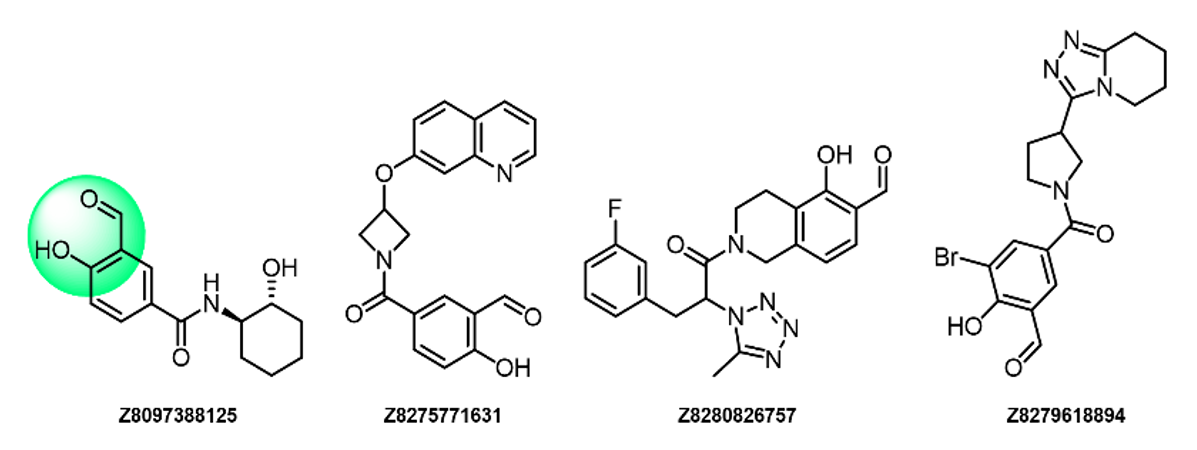
Examples of compounds in the library
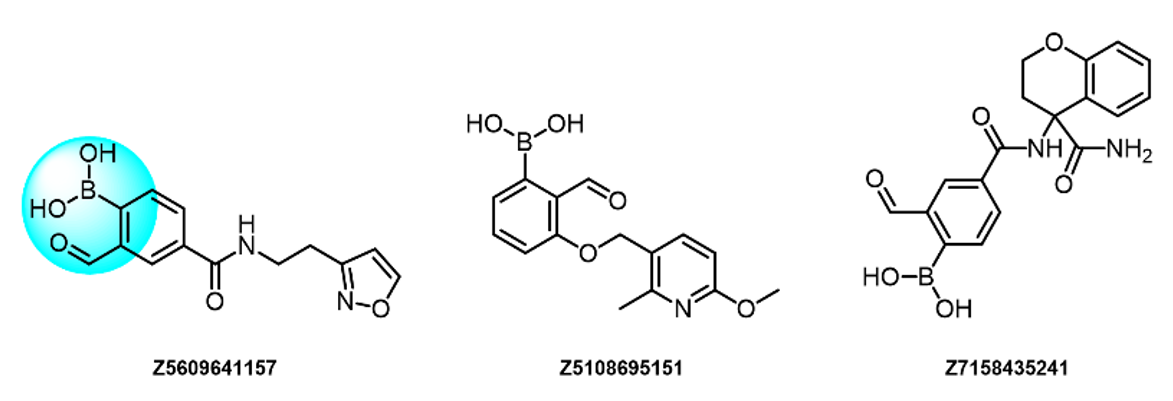
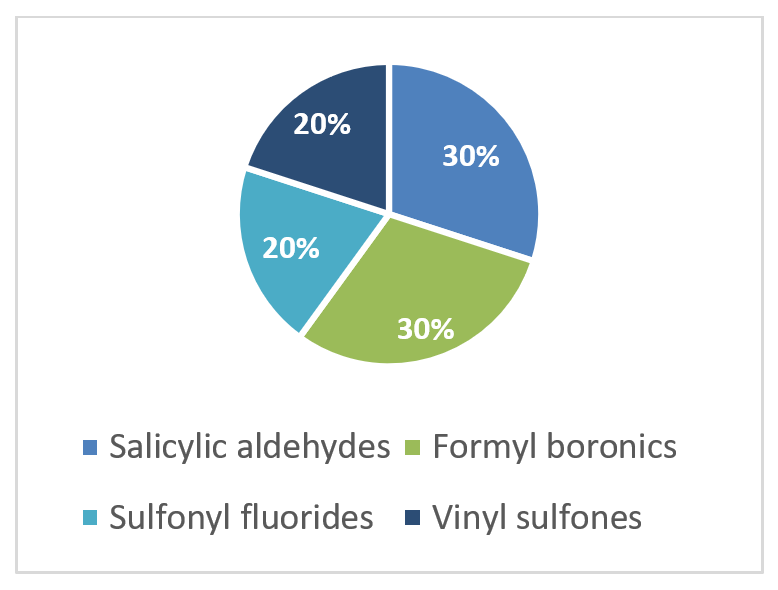
To address the increased interest and need for lysine-specific covalent binders, we have synthesized several libraries bearing warheads described as selective/preferred toward Lysine. Our renewed library contains only specially synthesized compounds with Lys-specific covalent warheads. The following types of covalent binders were used for the library construction:
Salicylic aldehydes
- Lys-specific, reversible
- The o-hydroxy group stabilizes covalent adducts
- Low reactivity
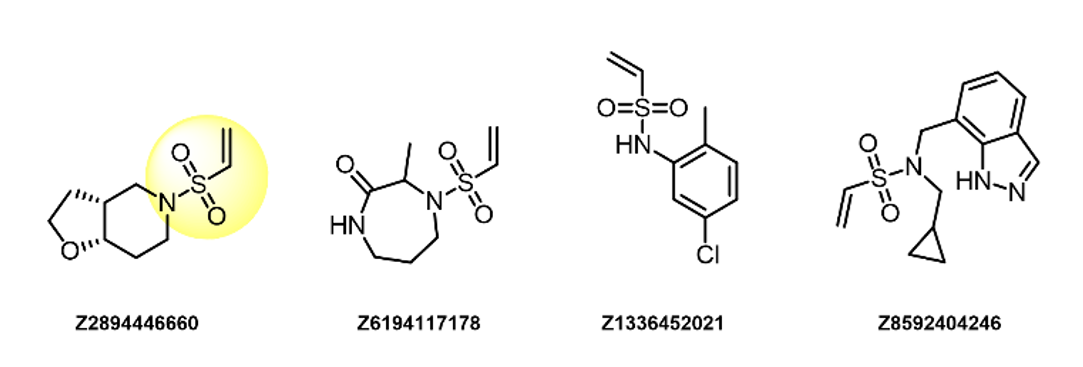
Vinyl Sulfonamides
- Irreversible binders
- Showed some selectivity toward Lys over Cys
- Highly reactive
α-Formyl Boronic acids
- Reversible and Lys specific
- Boronic residue in α-position dramatically enhances the stability of resulting adducts
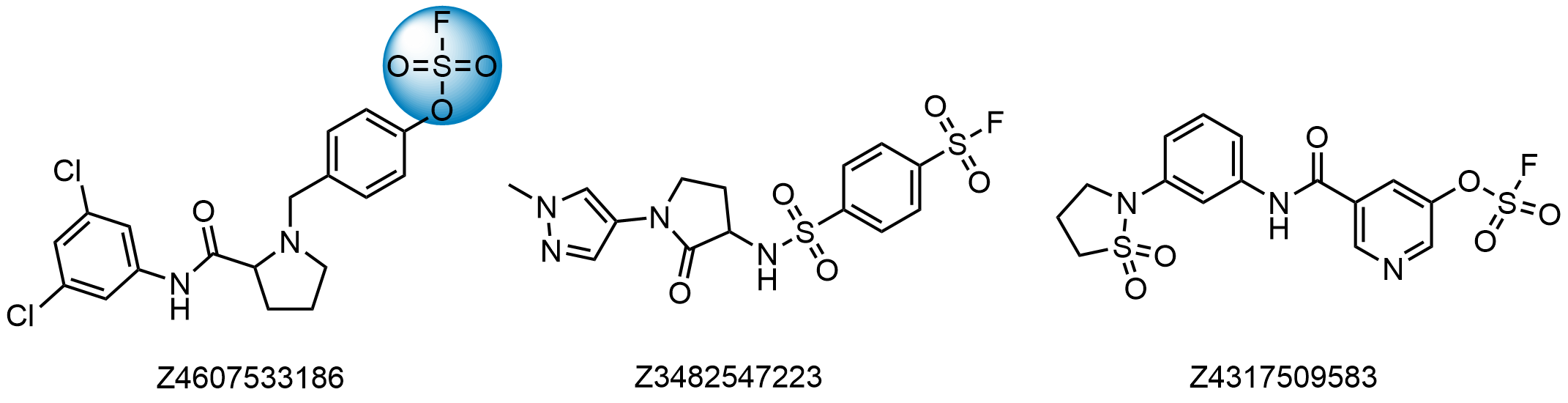
Sulfonyl Fluorides
- Irreversible binders for Lys, pH-dependent
- Reactivity depends on structure
- Compounds selected to display moderate reactivity
Distribution by covalent warhead
The library can be screened directly at Enamine in our biology laboratories, which offer a wide range of screening techniques including MS-based screening, kinetics, and intrinsic reactivity measurements. In this case, we will be happy to offer you a discount on library costs and follow-up services depending on the collaboration scope.
Ultimate tool for fragment screening
7 500 compounds
Solubility is critically important for fragment-based screening; assured solubility of fragments at high concentrations can prevent a number of issues during the screening procedure. We have confirmed experimentally aqueous solubility for 7 500 fragments in standard phosphate buffer at 1 mM; measurements were performed using nephelometry-based method. Representative subset of 3 000 compounds was designed using multi-vectoral diversity selection.
Key features:
- High structural diversity was achieved via two approaches: diversity selection using fingerprint-based Tanimoto distance and molecular framework frequency analysis. Compounds bearing trivial and abundant chemotypes were removed to enhance novelty of the set.
- Guaranteed aqueous solubility at 1 mM in PBS buffer and at 200 mM in DMSO
- Soluble Fragment Diversity Set can be readily followed with analogues either from stock or from validated syntheses. All required building blocks are available from Enamine stock.
Examples of the molecules in the library
Unique 3D diversity among shaped molecules
1 200 compounds
Enamine has been working on synthesis of new sp3-rich heterocycles and expanding of spirocyclic chemistry already over 16 years. Our own research in this area enabled synthesis of large variety of 3D shaped molecules that are well represented in stock screening collection. Synthesis of derivatized series of aliphatic and sterically hindered cores, often exclusive only for Enamine, led to a number of readily available analogs that is a crucial point in fragment hit optimization and follow-up stage.
Typical Formats
Catalog No.
3DF-1200-Y-10
Compounds
1 200
4 plates
Amount
10 µL of 100 mM DMSO stock solutions
Plates and formats
384-well microplates, Echo qualified Labcyte, 320 compounds per plate
Price
Catalog No.
3DF-1200-X-25
Compounds
1 200
15 plates
Amount
25 µL of 100 mM DMSO stock solutions
Plates and formats
96-well plates, Greiner Cat. No 650201, round (U) bottom, 1 & 12 columns empty, 80 compounds per plate
Price
Catalog No.
3DF-1200-X-50
Compounds
1 200
15 plates
Amount
50 µL of 100 mM DMSO stock solutions
Plates and formats
96-well plates, Greiner Cat. No 650201, round (U) bottom, 1 & 12 columns empty, 80 compounds per plate
Price
Download SD file
Library code: 3DF-1200
Version: 27 May 2021
1 200 compounds
at 100 mM in DMSO
Library design
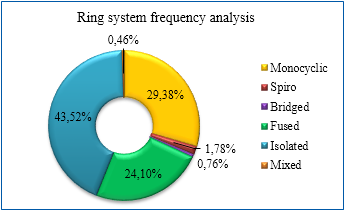
- Ro3 compliant dataset was refined with strict MedChem filters (FAF-Drugs3) and Fsp3cut-off 0.35
- 3D-dimensionality criteria: NPR1≥ 0.15; NPR2≥ 1.15 – value of npr1 (PMI plot 1)
- K-mean clustering of preselected 8 000 3D fragments has been carried out using NPR1/2 values. Only centroid molecules were included in the library, PMI plot 2
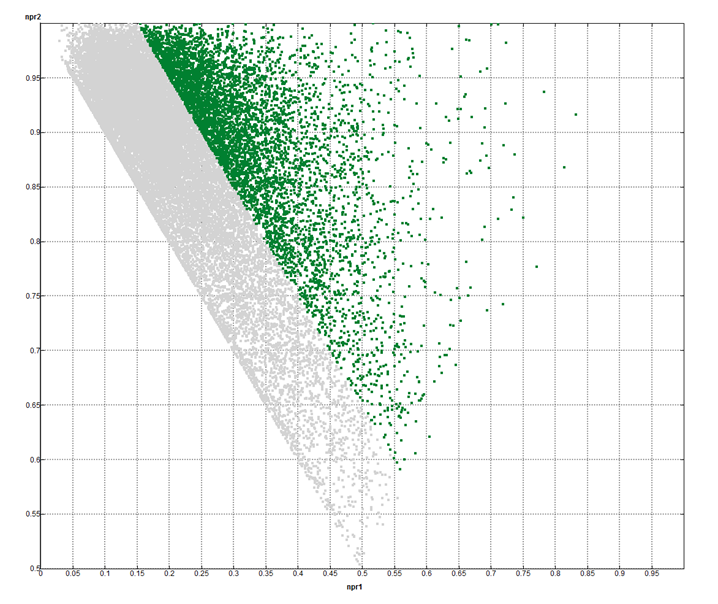
PMI plot 1: green dots correspond to 8 000 compounds indicated as 3D Fragments Set from Enamine in-stock Ro3 compliant molecules (grey dots).
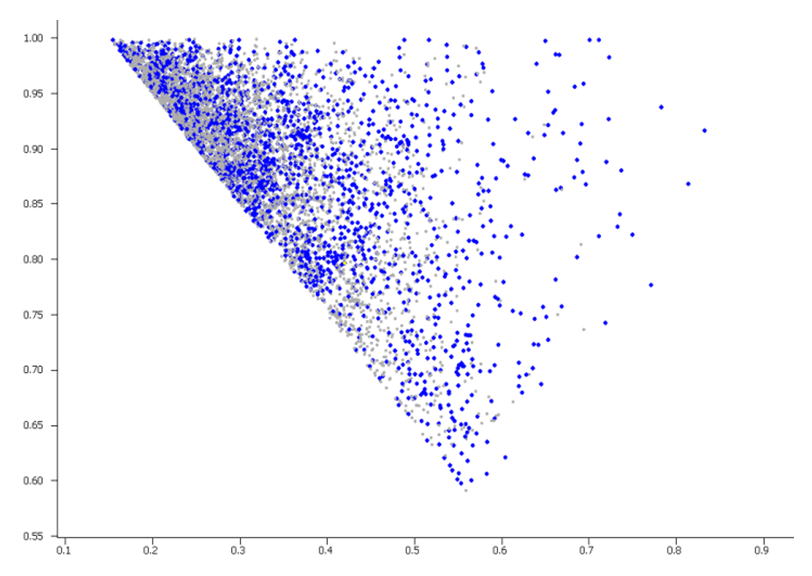
PMI plot 2: distribution of molecules in the library (centroids, blue spots) among of 8 000 of 3D-shaped initial
Examples of the molecules in the library
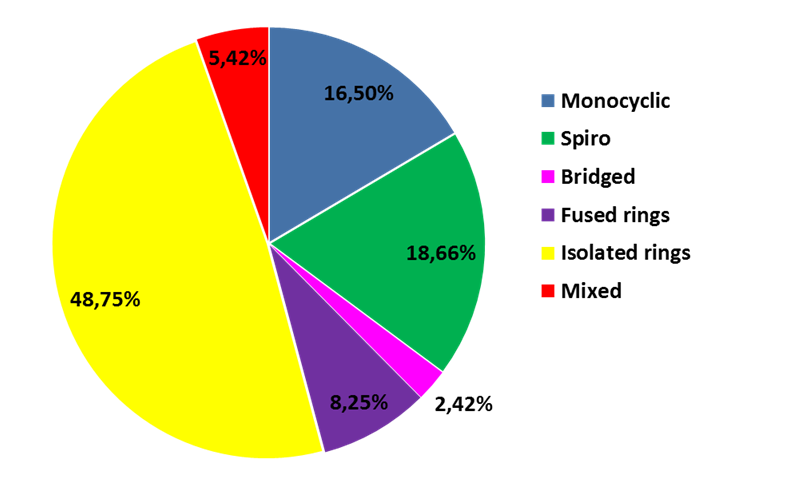
Distribution of ring systems among the library
Designed for discovery of novel hits in Immuno-Oncology therapeutic area
45 760 compounds
Immunotherapy has emerged as a transformative approach for cancer treatment being one of the most prospective fields in contemporary drug development. To address growing needs in novel and potent active molecules we designed special library focused on the most important targets: PD-1/PD-L1 checkpoints inhibitors, toll-like receptor family (TLR7 and TLR8, as a key players in antiviral response), IRAK4, ALK5 (one of JNK/P38 effectors and initiator of SMAD association), JAK-STAT pathway inhibitors, STING agonists, IDO inhibitors and the number of kinase targets – BTK, MAPK, VGFR and bRAF.
We have carefully selected 45 760 diverse compounds with the most promising features as potential ligands for known immune-oncology targets. All compounds are stored as dry materials and they can be acquired in diverse custom formats. Using our Immuno-Oncology Library for hit discovery you receive multiple benefits allowing you to save on time and costs in lead generation:
- Hit resupply and hit expansion from dry stock of over 4.7M compounds.
- Straightforward and low-cost analogs synthesis through our REAL Database technology.
- Fully customized hit-to-lead project support with broad capabilities available on-site.
You have also an option to screen the library directly at Enamine. We will be happy to offer you discount on library cost depending on the collaboration scope.
Download SD files
45 760 compounds for cherry-picking
Library design
In Immuno-Oncology Library design a special attention was paid to checkpoint proteins, such as PD-1, CTLA-4, CD152, CD279/74 and PD-L1.
In silico screening of different targets:
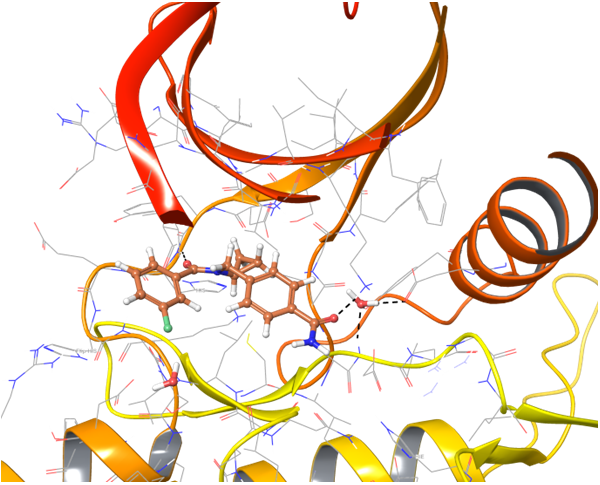
Structure-based approach was applied to search potential inhibitors of TLR7 and TLR8 receptors. All reported in PDB proteins structures were analyzed and superimposed for generation of protein structure-based pharmacophores. Thereafter two models have been validated with reference set of active and inactive compounds.
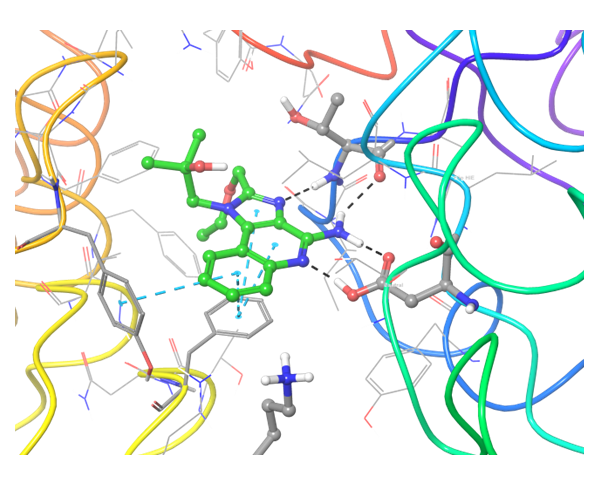
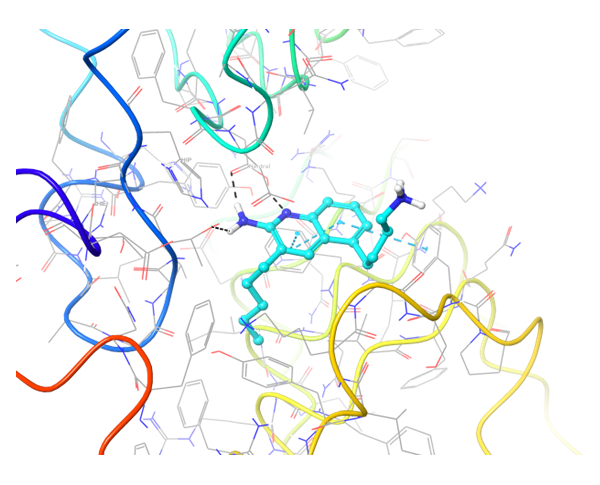
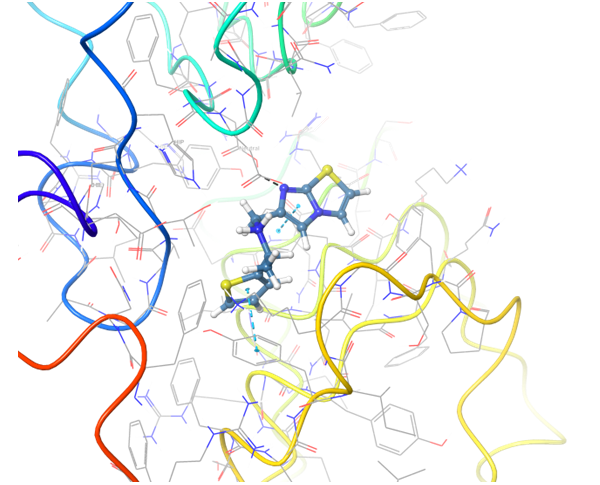
TLR7 and TLR8 in complex with co-crystallized ligands (upper pair). Binding mode of hit Z242112334 found after docking calculations with TLR7 (left) and another hit compound Z1438692506 in the cavity of TLR8 (right).
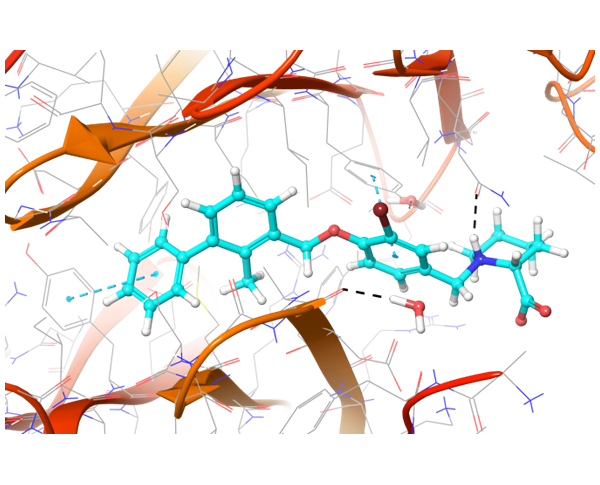
Two types of transmembrane proteins (IRAK4 and TGF beta R1) with reported kinase activity were studied based on 4U97 and 2WOT structures respectively. The presence of several hydrophobic cores in the ligand structure and its ability to form strong h-bonds with key amino acids in the binding site were requirements for potential inhibitors selection. However, to overlap all possible structures and conformations an alternate combination of hydrophobic cores and h-bonds was used for selection of compounds in these mini libraries. Two subsets of constraints were generated to create the most exhaustive screening model for each kinase.
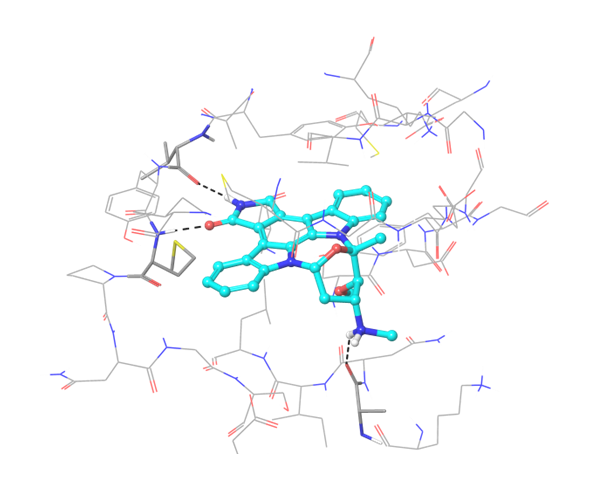
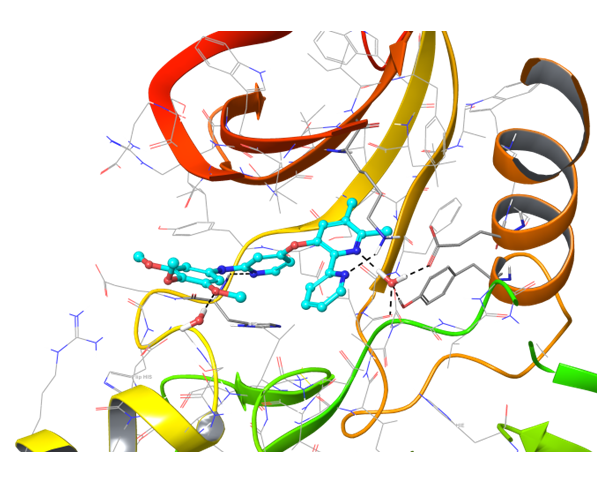
IRAK4 in its crystalized form and in complex with docked Z243559212 are in the left column. TGF beta R1 in original state from RCSB and in complex with Z316882020 are in the right column.
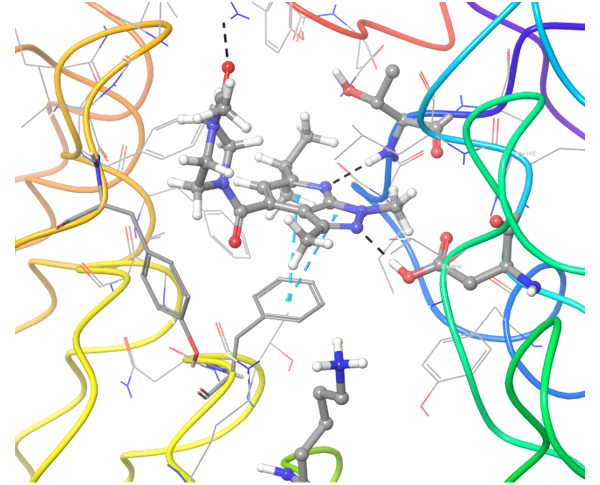
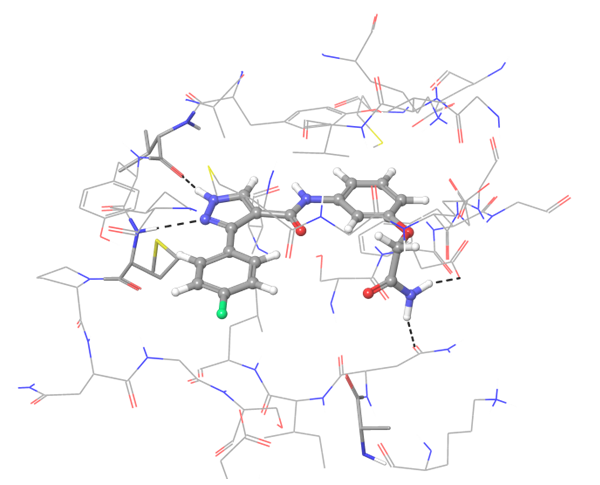
In case of PD-1 and PD-L1 binding, there are few known PD-L1 inhibitors. We applied structural and spatial constraints to find compounds, which can similarly modulate PD-L1 structure. To increase selectivity and at the same time avoid the conformational similarity, two-staged screening was performed, resulting in the selection of the most potent binders.
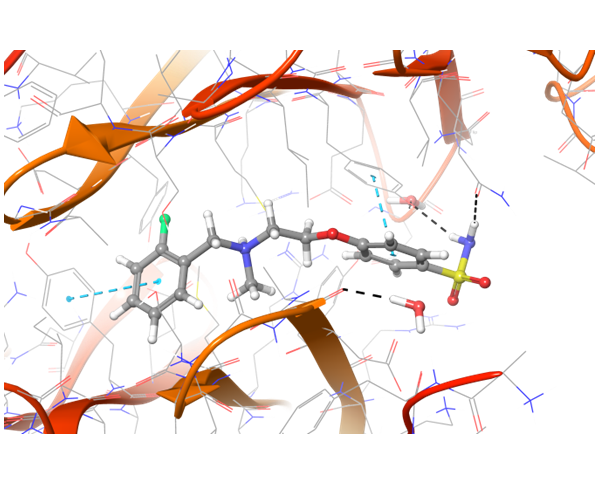
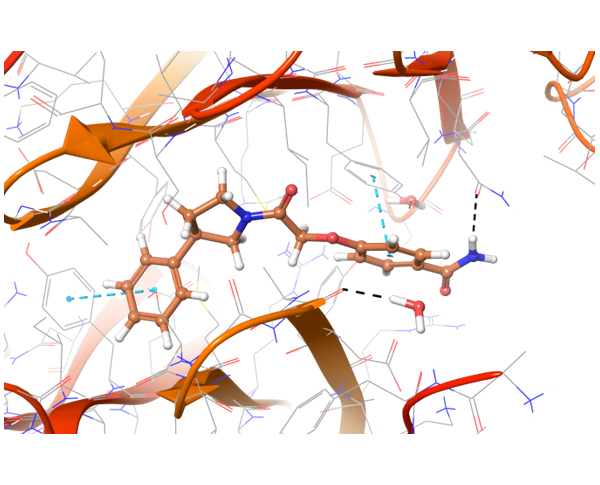
Co-crystallized BMS-8 structure (upper left) with indicated interactions and both Z195611316 and Z942299126 ligands, which have similar structure and binding modes.
Targeted Libraries
Product catalog
AGR-10
Size
10 240
compounds
Description
Library of compounds intended for use in agro/crop science
Download file
AGR-14
Size
14 160
compounds
Description
Designed for discovery of novel allosteric ligands
Download file
Size
4 800
compounds
Description
Carefully selected molecules via docking and visual evaluation
Download file
ABAC-32
Size
32 000
compounds
Description
Designed for the discovery of novel antibacterials
Download file
ATB-2500
Size
2 500
compounds
Description
Designed for the discovery of new effective and safe treatment
Download file
AVR-3200
Size
3 200
compounds
Description
Designed for discovery of new Nucleoside-like antivirals
Download file
Size
1 348
compounds
Description
Designed for discovery of new water channels modifiers
Download file
Size
7 171
compounds
Description
Designed for discovery of novel BACE inhibitors
Download file
BRD-15
Size
15 360
compounds
Description
Specially selected molecules to target bromodomains
Download file
CICL-10560
Size
10 560
compounds
Description
Designed for discovery of new Voltage-gated calcium channel blockers
Download file
CNS-47
Size
47 360
compounds
Description
Library of novel small molecules with high CNS MPO scores
Download file
CNSd-5
Size
5 440
compounds
Description
Sublibrary of CNS-47 Library
Download file
COV-16800
Size
16 800
compounds
Description
Designed for the discovery of new SARS-CoV-2 and pan-Coronavirus antivirals
Download file
DNA-5760
Size
5 760
compounds
Description
Designed for identification of new actives against proteins essential for DNA stability
Download file
EPG-38080
Size
38 080
compounds
Description
Library of compounds focusing to hit on a number of epigenetic targets
Download file
ERL-8960
Size
8 960
compounds
Description
Designed to effectively target the receptor and block estrogen release
Download file
GML-2470
Size
2 470
compounds
Description
Specially synthesized set of compounds able to mimic glycosides and their interaction with proteins
Download file
GPR-53
Size
53 440
compounds
Description
Designed for discovery of new GPCR ligands
Download file
HBL-24
Size
24 000
compounds
Description
Designed for discovery of novel kinase ATP pocket binders
Download file
IDO-4800
Size
4 800
compounds
Description
IDO focused library designed by a combination of structure- and ligand-based methods
Download file
Size
45 760
compounds
Description
Designed for discovery of novel hits in Immuno-Oncology therapeutic area
Download file
ICL-36
Size
36 800
compounds
Description
Designed for discovery of new Ion Channels ligands
Download file
JAK-STAT-1280
Size
1 280
compounds
Description
Designed for efficient hit finding against a number of immune disorders, including RA
Download file
KNS-64960
Size
64 960
compounds
Description
Designed for discovery of novel protein kinase inhibitors
Download file
KYN-13
Size
13 120
compounds
Description
Designed for discovery of new regulators of methabolic disorders
Download file
LGR-6400
Size
6 400
compounds
Description
A sub-library of Enamine’s GPCR Library designed for discovery of novel lipid GPCR ligands
Download file
Size
1 388
compounds
Description
A set of LOXs inhibitors designed using docking and 2D similarity search
Download file
Size
2 468
compounds
Description
A set of compounds focused on targeting molecular chaperones
Download file
NML-320
Size
320
compounds
Descriptions
Small library of specially synthesized compounds
Download file
PDZ Domain Library
PDZ-1920
Size
1 920
compounds
Descriptions
Sublibrary of PPI-40
Download file
PML-8960
Size
8 960
compounds
Descriptions
Selected molecules able to mimic common protein motifs
Download file
Size
40 640
compounds
Descriptions
Designed for discovery of novel PPI inhibitors
Download file
RNA-28
Size
28 000
compounds
Descriptions
Designed to promote the discovery of new-generation medicines
Download file
CSHL-12160
Size
12 160
compounds
Descriptions
Designed for discovery of mild electrophilic inhibitors of the largest enzyme class
Download file
SICL-5440
Size
5 440
compounds
Descriptions
Designed for discovery of new Nav1.7 channel blockers
Download file
TBL-3200
Size
3 200
compounds
Description
Library of potential tubulins ligands
Download file
WPL-10
Size
10 560
compounds
Description
Designed for the discovery of new effective modulators of Wnt/β-catenin signaling pathway
Download file
Support
We offer comprehensive support in developing your hit compounds. Naturally such programs are realised most efficiently when biological actives originate from our screening collection. However, even if the hit compounds are from the collections of other vendors lead identification and optimization projects can proceed most productively in our hands. Sometimes for this we only need to synthesize first examples of the given chemical series and validate synthesis route.