Designed for discovery of novel BACE inhibitors
7 171 compounds
Alzheimer disease (AD) is one of the most difficult to treat CNS disorder. Despite of intensive investigation over last decades no effective medication has been developed against this horrible disease. However, the recent data on beta-secretase inhibition showed budding results in this field. BACE inhibition would seem to be a powerful tool against AD evolution in the early stages of the diseases. These data served as a basis for design of high-quality BACE targeted library with most diverse chemotypes and good CNS profile.
Download SD files
7 171 compounds for cherry-picking
Library Design
The database of activities related to BACE was carefully combined (as a result of searching in ChEMBL and literature data). In total 7 465 compounds with reported sufficient activity were found. All available PDB structures were analyzed as well. 23 most representative PDB complexes were used for molecular docking calculations. Further algorithm to search potential regulators of BACE target is presented below:
By using PDB, PDBe, pfam all available structures were selected. Also based on literature data and ChEMBL all active compounds were selected.
23 most representative structures were selected for molecular docking (2b8v, 2ohr, 2viz, 2wjo, 2zdz, 3exo, 3fkt, 3hw1, 3in3, 3l38, 3l5b, 3l5f, 3rsx, 3udy, 3uqw, 3vf3, 4acu, 4i10, 4l7h, 4xkx, 5hdv, 5kqf, 5uyu).
Then using obtained structures for BACE receptors and Enamine chemical DataBase flexible molecular docking.
As result for BACE protein target 7 171 (docking library) compounds were selected.
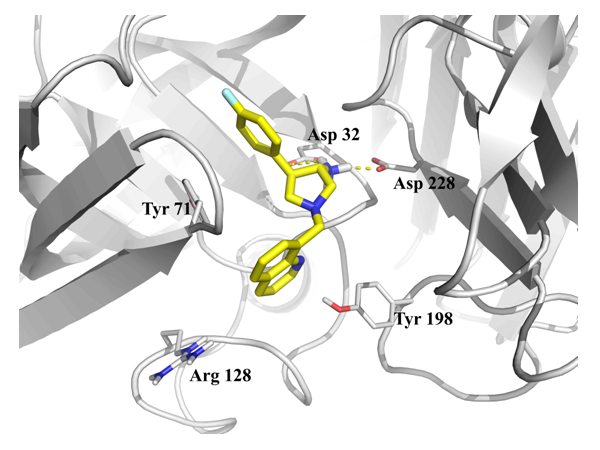
Fig. 1.Example of result of docking (4l7h).
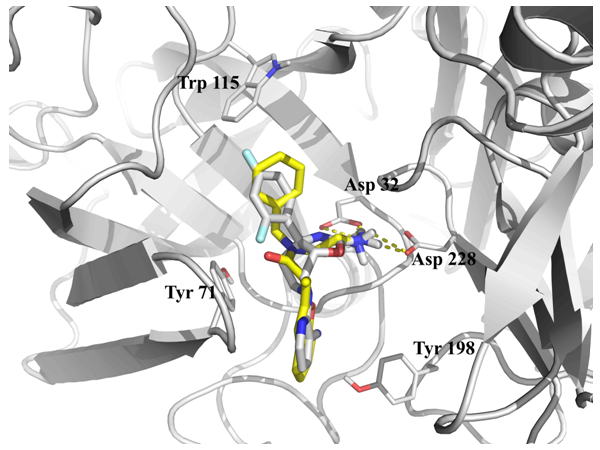
Fig. 2. Binding of 2ohr with native ligand (gray) and result of docking (Yellow).



