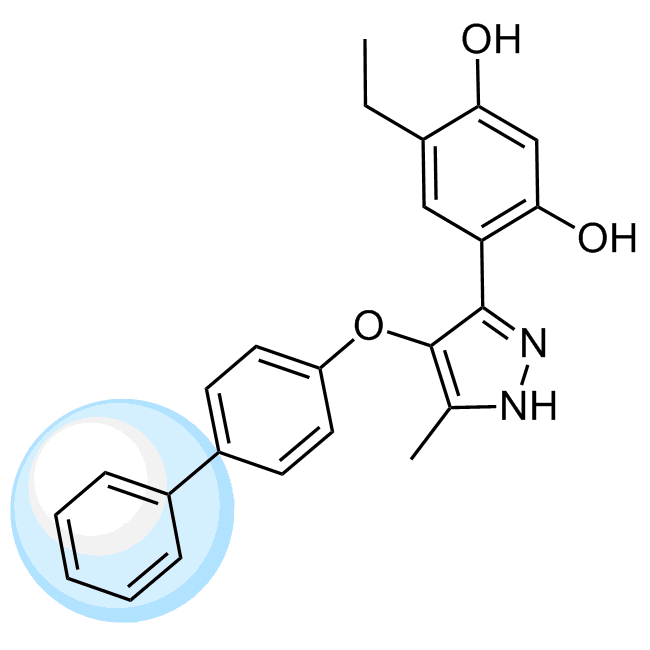
A set of LOXs inhibitors designed using docking and 2D similarity search
1 388 compounds
Lipoxygenases (LOXs) are enzymes that catalyze formation of the corresponding hydroperoxides from polyunsaturated fatty acids (e. g. linoleic or arachidonic). LOXs are widely expressed in immune, epithelial, and tumor cells, and activation of these enzymes induces numerous structural and metabolic changes. Abnormal LOX activity contributes to a number of pathophysiological conditions including inflammation, skin disorders and tumor genesis.
The 1 388 - compound Enamine lipoxygenase library was designed using two different approaches:
- Docking-based in silico screening, which identified compounds derived from novel scaffolds;
- Similarity search using 2D linear fingerprints and 3D pharmacophore features of the reported lipoxygenase inhibitors, which gave molecules with new side chains.
Download SD files
1 388 compounds for cherry-picking
Library design
Molecular docking
Docking models were built from the PDB reported protein structures 3O8Y, 4NRE and 3D3L. Prior to docking, all structures were optimized and reconstructed to correct gaps and missing side chains (Figure 1). Water molecules coordinated to Fe3+ ion were restrained, and their ionization state was assigned. Each docking model contained electrostatic descriptors of the binding site and mandatory positional constraints.
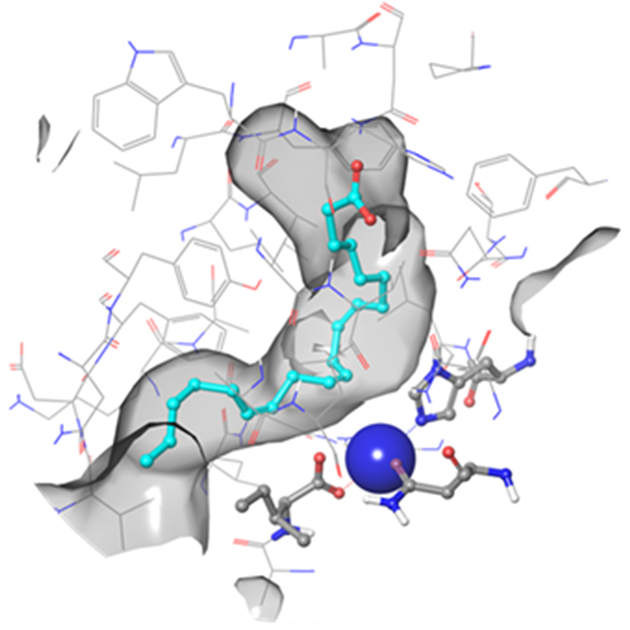
Fig. 1. hLOX-5 binding site in a surface volume representation (channel mode) with a native substrate –arachidonic acid (Fe3+ ion is shown in blue)
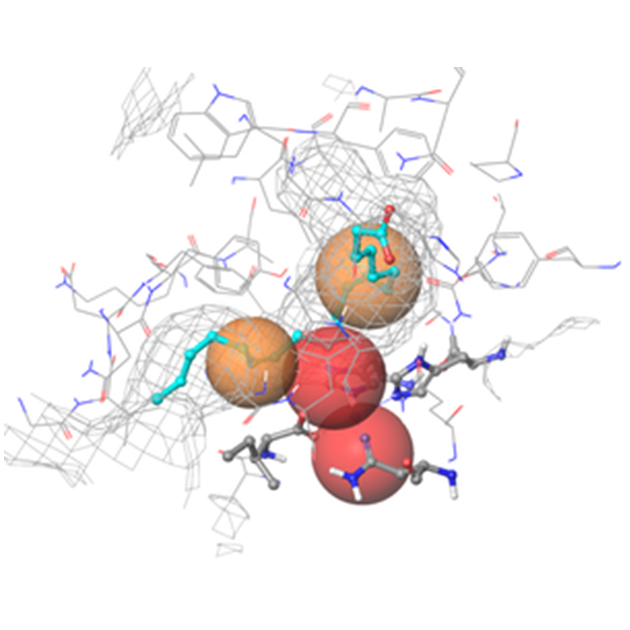
Fig. 2. Binding site of hLOX-15-II (4NRE) in grid representation with the most important pharmacophore features indicated: volume/shape, hydrophobic areas (colored in brown), H-bond acceptors and electron deficient fields (colored in red)
The resulting models were used to screen the MedChem refined subset of ~1.0M molecules of Enamine’s entire stock collection (over 4.7M compounds). Hits were defined as possessing an obligatory alignment for the positional constraints – metal coordination (red) and hydrophobic center (orange) (Figure 2). The docking results were evaluated through comparison of the docking scores and visual inspection of binding poses (presence of H-bonds, degree of exposure to water), which gave 950 compounds (Figure 3).
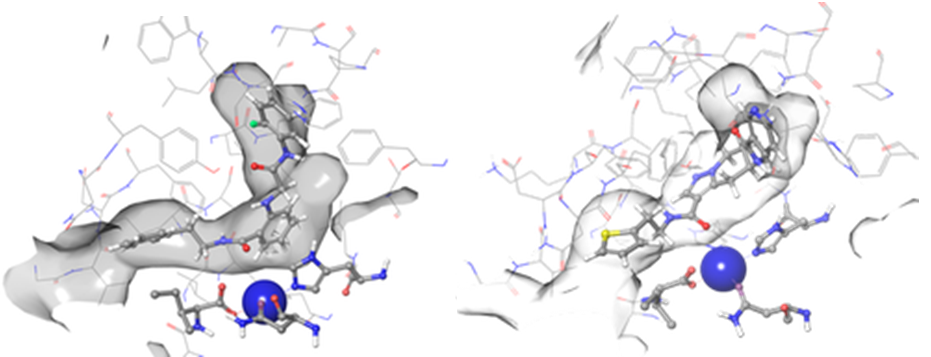
Figure 3. Examples of hit binding poses with high scoring functions identified after docking – Z31004492 (left) and Z1849138647 (right). Both molecules form coordination bonds with Fe3+ ion (shown in blue) and match two hydrophobic features in the binding pockets
Similarity search
- Search of ChEMBL database against the lipoxygenase targets gave a reference set of 322 active compounds for three lipoxygenase types (A5, A12, A15). These compounds were further filtered using potency values cut-off (IC50 ≤ 1μM) and MedChem toxicophore filters.
- 2D similarity search with a 0.85 threshold was done using linear fingerprints and Tanimoto algorithm.
- Nearly 500 compounds were selected from Enamine’s screening collection and added to the LOX library.
Examples of the molecules in the library
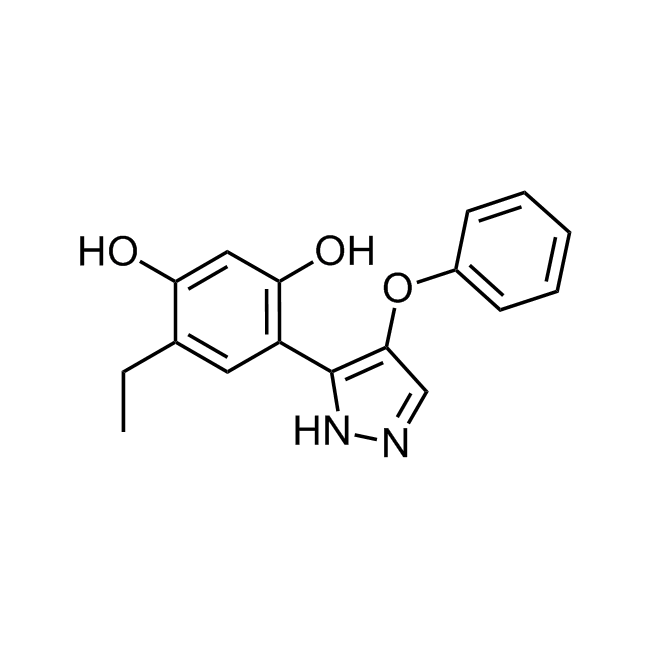
CHEMBL381149 IC50= 920 nM, 15-hLOX
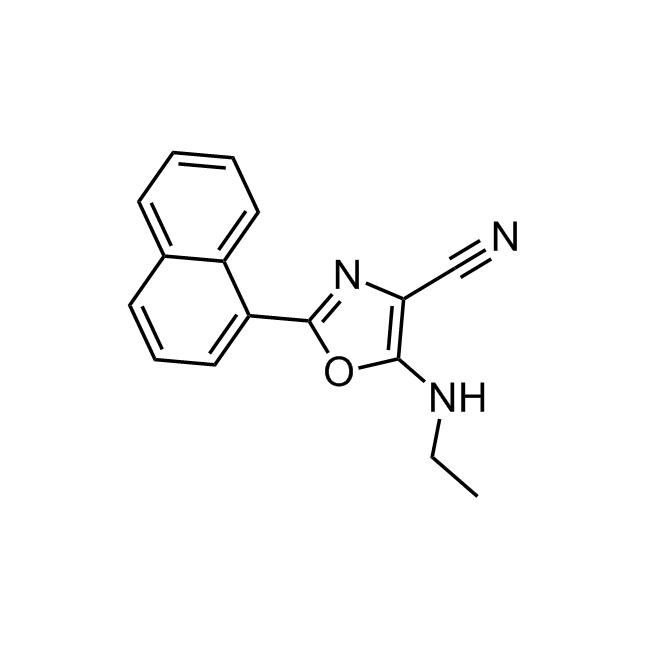
CHEMBL1571456 IC50= 120 nM, 15-hLOX
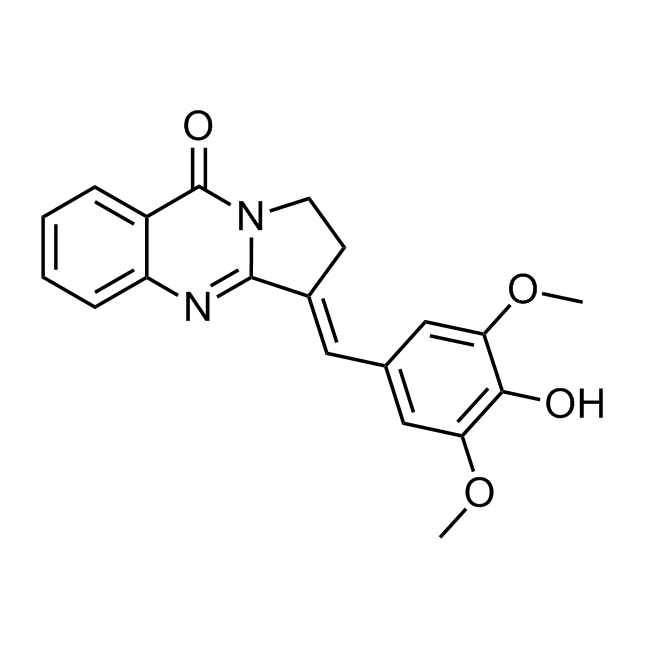
CHEMBL518103 IC50= 40 nM, 5-AhLOX