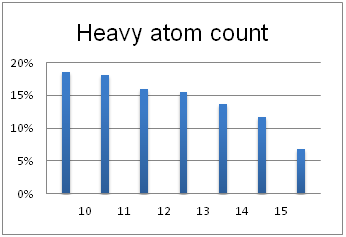
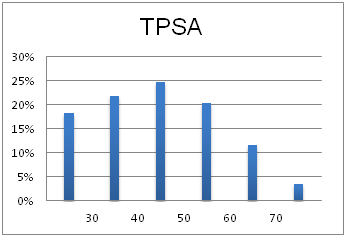
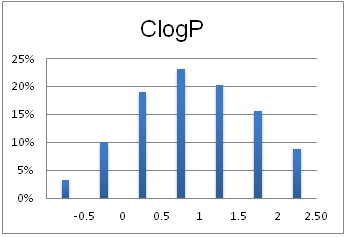
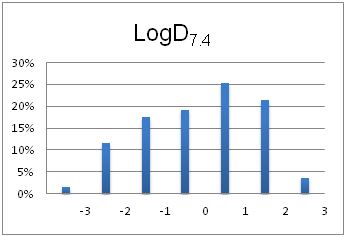
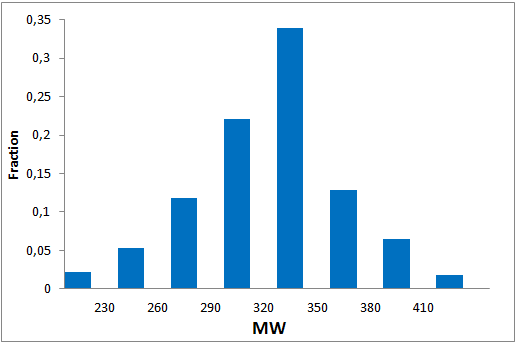
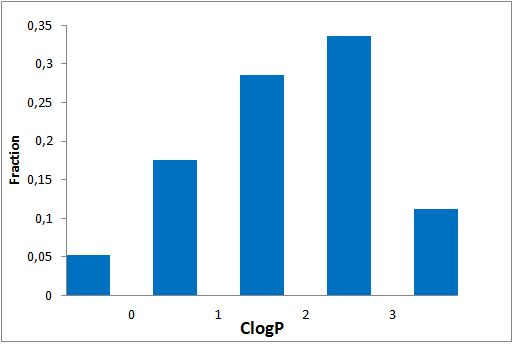
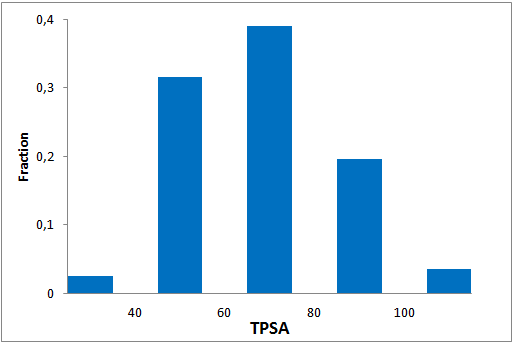
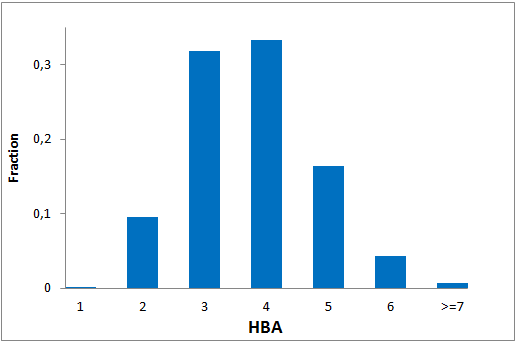
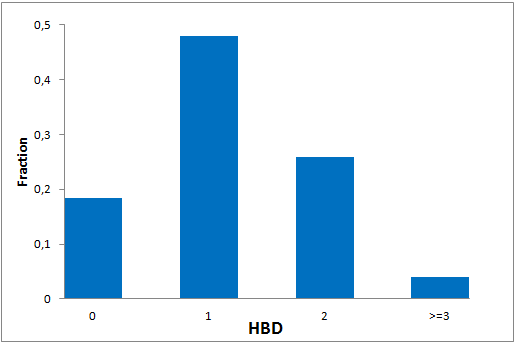
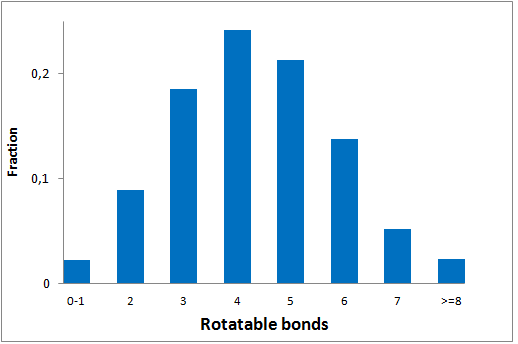
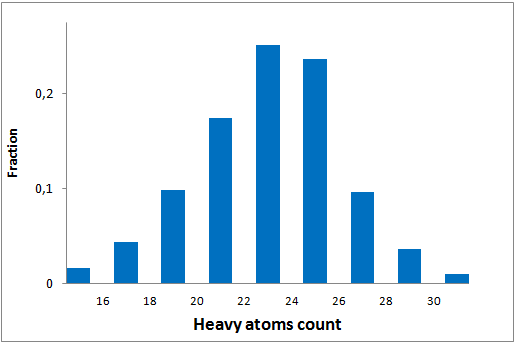
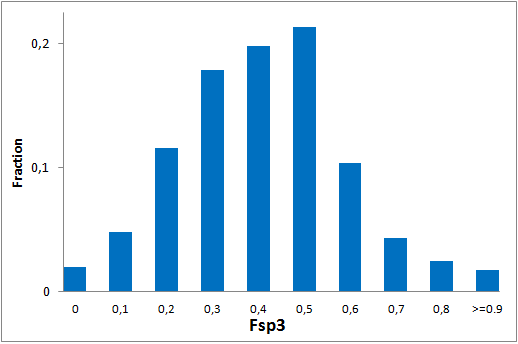
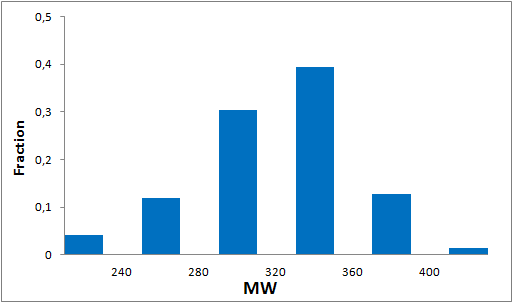
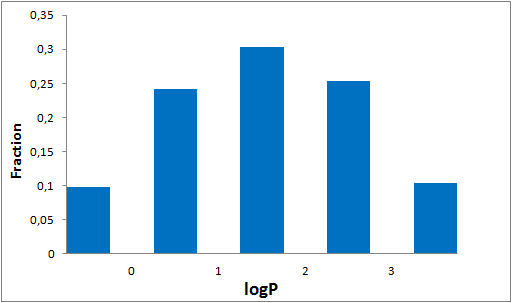
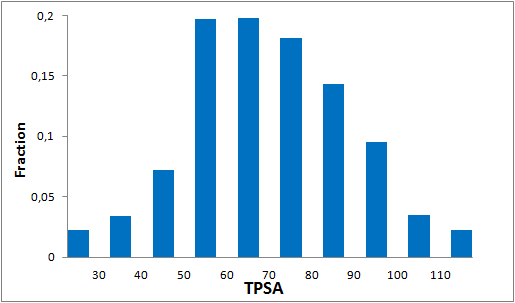
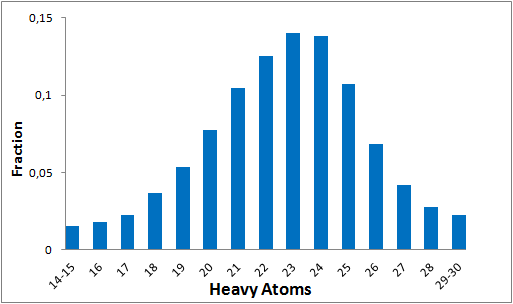
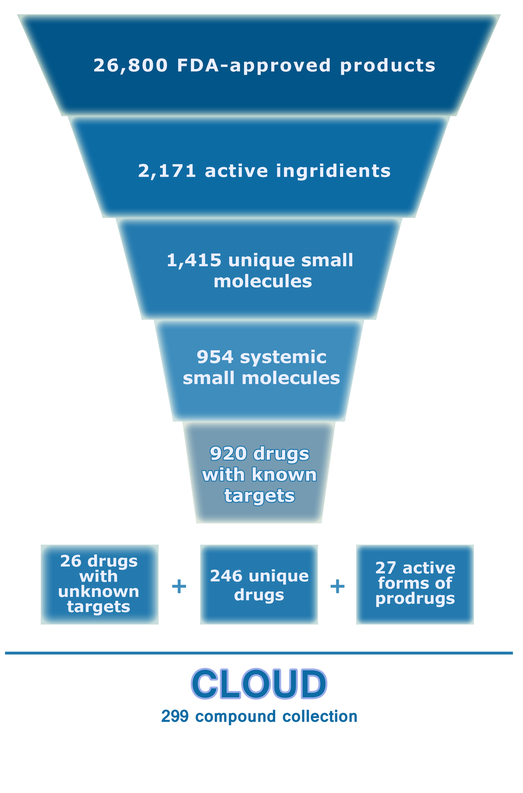
The comprehensive Drug Collection
299 compounds
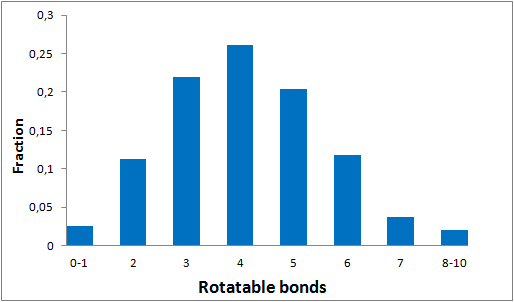
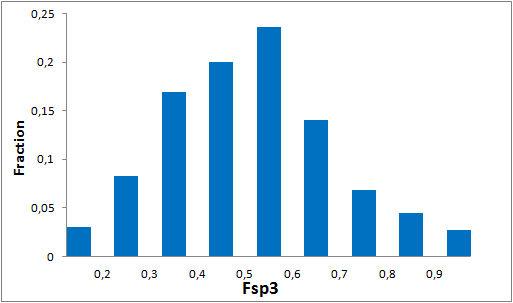
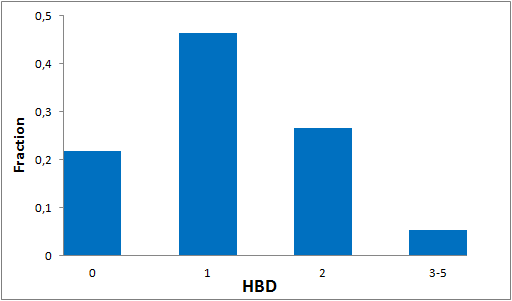
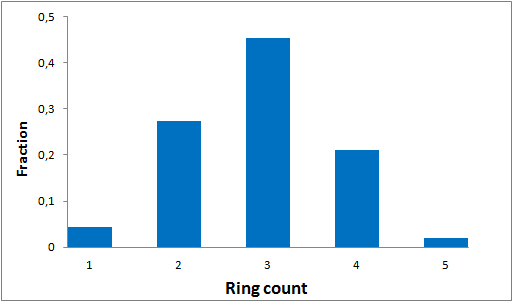
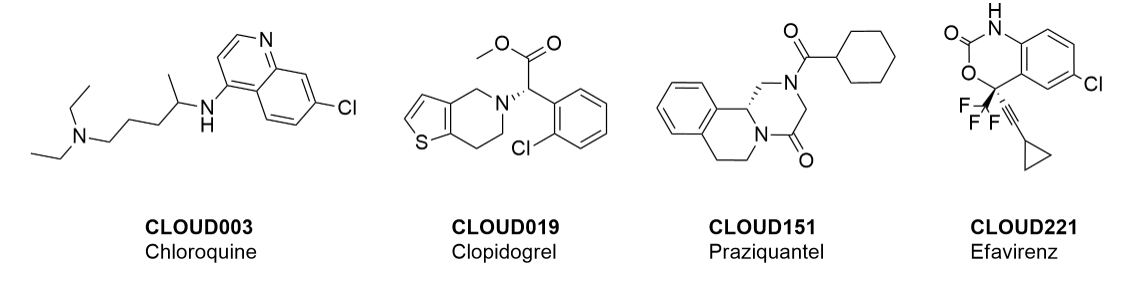
To date no commercially available compound collection covered all drug classes, resulting in complications in combinatorial screening attempts. We collaborated with a group of researchers at CeMM and collected a condensed screening set of 299 small molecules representing the entire target and chemical space of all FDA-approved drugs. The CeMM Library of Unique Drugs (CLOUD) covers prodrugs and active forms at pharmacologically relevant concentrations and is ideally suited for combinatorial studies. Recent publication [1] provides evidences of successful drug repurposing by pairwise compound screening from CLOUD collection in cancer cell viability assay. The findings highlight that refined chemical library CLOUD is a powerful tool for drug repurposing and evaluation of new substances suitable for all high-content and high-throughput assays.
CLOUD is now available for immediate supply in the following most popular pre-plated formats
Typical Formats
Catalog No.
CLOUD-Y-0
Compounds
299
Amount
≤ 1 µL of 10 mM of DMSO solutions
Plates and formats
384-well echo-qualified assay ready microplates, Greiner/Labcyte
Price
Catalog No.
CLOUD-Y/X-10
Compounds
299
Amount
10 µL of 10 mM of DMSO solutions
Plates and formats
384-well or 96-well plates, echo-compatible
Price
Catalog No.
CLOUD-Y/X-50
Compounds
299
Amount
50 µL of 10 mM of DMSO solutions
Plates and formats
384-well or 96-well plates, echo-compatible
Price
Download SD files
Key features
- Covers all therapeutically significant space of approved drugs.
- Contains prodrugs and active metabolites at pharmacologically relevant concentrations.
- Ideally applicable for combinatorial assays to study new drug combinations.
- Most structurally diverse drug library among commercially available collections.
- Immediately accessible in convenient pre-plated formats with carefully prepared detailed documentation. The library can be also made in any customized ready-to-screen formats.
Examples of drugs in the library
Selection and library evolution scheme

[1] Nature Chemical Biology 13, 771–778 (2017).
Fragments of high MedChem tractability
1 920 compounds
Fragment screening usually provides high numbers of hits and is one of the most affordable approaches to start drug discovery. However, subsequent steps involving hit-to-lead optimization may be challenging and require significant resources. It’s even more painful when efforts result in false positives.
We have designed a new fragment library specifically addressing the quality of decision making when evaluating hits. Our High Fidelity Fragment Library features high medchem tractability enabling researchers to grow interesting hits with confidence.
High medchem tractability of this set was achieved through structure review and selection by FBDD experts at Takeda and Carmot Therapeutics. In particular, we would like to thank Dr. Derek Cole, Dr. Dan Erlanson, Dr. David Lawson and Dr. Xiaolun Wang for their involvement in the design of our High Fidelity Fragment Library.
High quality: All compounds selected for this library passed turbidity tests to assure high solubility in water at 1 mM; all aggregators were filtered out. In addition, the fragments in this set were screened by surface plasmon resonance (SPR) to remove any false positive fragments. SPR screening was kindly provided by Dr. Delphine Collin at HarkerBio.
Optimal molecular properties: Fragments in this set all have 9–16 heavy atoms, are moderately complex and have suitable physiochemical and shape profiles.
Our High Fidelity Fragment Library is available for prompt delivery in various formats. The most popular library options are listed below:
Typical Formats
Catalog No.
HFF-1920-Y-10
Compounds
1 920
6 plates
Amount
10 µL of 100 mM DMSO stock solutions
Plates and formats
384-well microplates, Echo qualified Labcyte LP0200
Price
Catalog No.
HFF-1920-Y-25
Compounds
1 920
6 plates
Amount
25 µL of 100 mM DMSO stock solutions
Plates and formats
384-well microplates, Echo qualified Labcyte PP0200
Price
Catalog No.
HFF-1920-X-50
Compounds
1 920
24 plates
Amount
50 µL of 100 mM DMSO stock solutions
Plates and formats
96-well plates, Greiner, first and last columns empty
Price
Download SD file
Library code: HFF-1920
Version: 14 November 2024
1 920 compounds
at 100 mM in DMSO
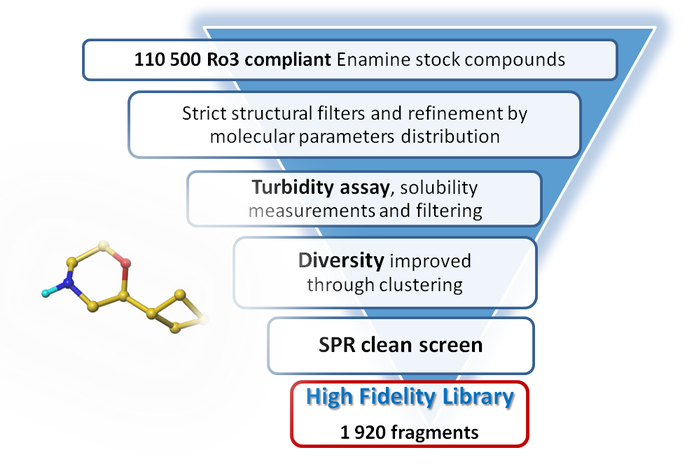
Library design
A complex iterative approach has been applied in the design of this library. Fragment selection was made from over 260k Ro3-compliant compounds in Enamine’s stock collection. All industry recommended medchem filters, including PAINS, were applied. Diversity selection was performed using clustering algorithm and manual review of identified clusters to select the most representative structure within each cluster.
All compounds were tested for solubility in water and aggregation by laser nephelometry to remove compounds with solubility issues. The resulting set of fragments was then tested in clean SPR screens to exclude SPR “sticky” compounds.
The selection process and resulting molecular parameters of our High Fidelity Fragment Set are described in the following scheme:
Designed for efficient hit finding against a number of immune disorders, including RA
1 280 compounds
The library was designed to be a universal tool to search potential JAK-STAT pathway modulators. Protein structure-based analysis and scaffold-hopping approach were used to create an optimal in silico screening models. Additionally, ligand-based approach has been applied to enrich the library with topological analogs and similar compounds to reported actives.
Resulted library is as a generic starting point for ligand search, regardless of particular SH2 domain of interest, with high probability of initial hit discovery. The library was validated with in vitro screening against SRC SH2 domain resulting in 3.5 % of identified hits.
Download SD files
Library design
All available structures of SH2 domain binding sites were analyzed and clustered based on their spatial molecular shape. After the alignment analysis and clustering structures with most different conformations were selected for virtual screening. Enamine MedChem filtered in-stock subset (~1 M compounds) with additional selection of compounds carrying peptidomimetic motifs was used for molecular docking calculations.
Multiple sequence alignment was applied to 120 SH2 domains contained within 110 proteins. Over 200 protein structures were extracted from Protein Data Bank (PDB): 66 NMR-based structures and 153 derived from X-ray crystallography experiments. As variation of the binding site conformation may significantly influence its binding properties, all files were split into individual structures, resulting in total structure count of 1633.
Key pharmacophore interaction points: pTyr binding pocket, carbonyl O in the binding site center, hydrophobic sub-pocket.
Solvent accessible molecular surface within 12 Å from the binding site center was used for calculation of shape-based numeric descriptors. 3D structures were clustered based on 3D shape similarity. 8 spatially diverse structures were selected for docking.
Table 1. Summary of protein 3D structures clustering results and structures selected for docking.
1
Centroid structure (PDB id, chain, NMR model)
1o49, chain A
Centroid structure: organism, gene, domain
Homo Sapiens SRC SH2
Structures in cluster
223
2
Centroid structure (PDB id, chain, NMR model)
2fci, chain A, model 6
Centroid structure: organism, gene, domain
Bos Taurus PLCG1 SH2-2
Structures in cluster
48
3
Centroid structure (PDB id, chain, NMR model)
2ge9, chain A, model 15
Centroid structure: organism, gene, domain
Homo Sapiens BTK SH2
Structures in cluster
72
4
Centroid structure (PDB id, chain, NMR model)
3in7, chain A
Centroid structure: organism, gene, domain
Homo Sapiens GRB2 SH2
Structures in cluster
183
5
Centroid structure (PDB id, chain, NMR model)
2jyq, chain A, model 9
Centroid structure: organism, gene, domain
Homo Sapiens GRB2 SH2
Structures in cluster
114
6
Centroid structure (PDB id, chain, NMR model)
2k7a, chain B, model 5
Centroid structure: organism, gene, domain
Mus Musculus ITK SH2
Structures in cluster
218
7
Centroid structure (PDB id, chain, NMR model)
2kk6, chain A, model 14
Centroid structure: organism, gene, domain
Homo Sapiens FER SH2
Structures in cluster
149
8
Centroid structure (PDB id, chain, NMR model)
1uus, chain A
Centroid structure: organism, gene, domain
Dictyostelium Discoideum DSTA SH2
Structures in cluster
129
Virtual screening against Stat3beta:
- Collaborator: Gyeong Baeg, NYMC
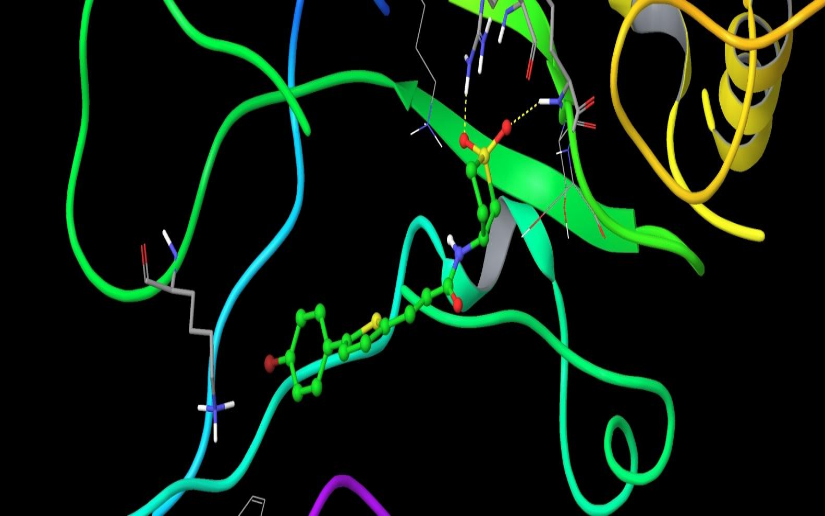
Chemical compounds are significantly smaller in size than the natural peptide interacting with this target. Therefore, when creating the library, we tried to fill all potential sub-pockets available near the phosphotyrosine binding site. The entire available surface of the protein was conditionally broken down into 5 models and proceeding from this was carried out post-docking analysis. As an example, the first two models:
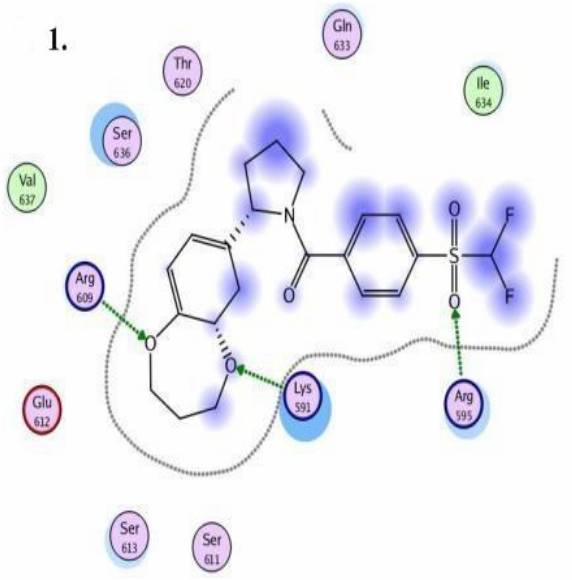
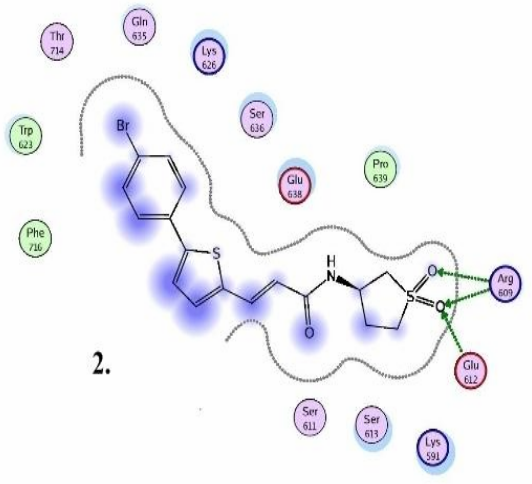
STAT3 models: Schematic representation of ligand binding pocket
The largest diversity library with high MedChem tractability
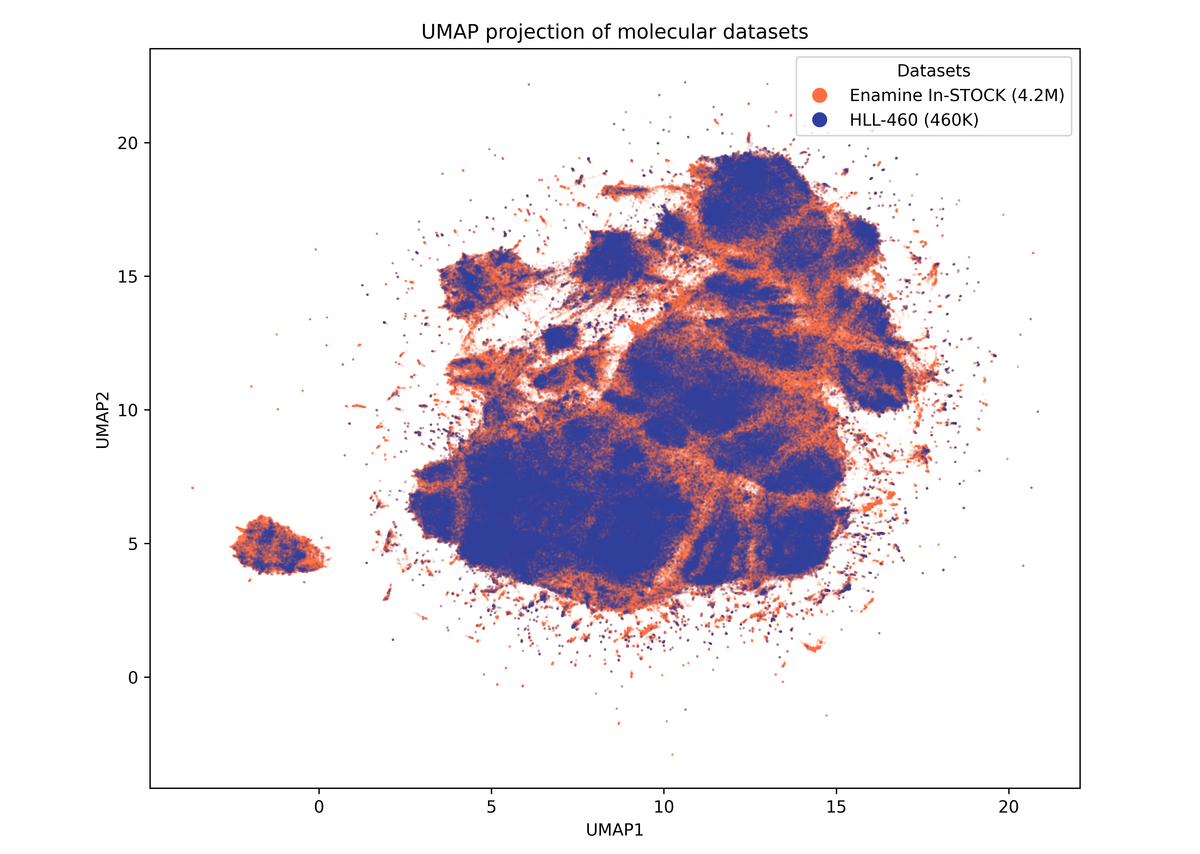
460 160 compounds
Hit Locator Library (HLL) represents the entire Enamine’s Screening Collection of 4.7M+ compounds. Smart clustering approach and elaborated MedChem filters together with thoughtful molecular parameters restriction allowed us to design an ultimate HTS Library.
Typical Formats
Hit Locator Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
HLL‑460-0-Z-10
Compounds
460 160
360 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
HLL‑200-10-Y-10
Compounds
200 000
625 plates
Amount
10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
HLL‑300-50-Y-10
Compounds
300 160
938 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
HLL‑460-0-Z-10 screening library 460 160 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
5 120 compounds
Library design
Carefully considered approach to the design our largest screening library aims to deliver the most reliable source of initial hits. Each compound in the library passed stringent MedChem filters and QC control. Construction of this HTS Library allow to get initial SAR data immediately after a first screen and will help you to determine safe hits, which you can follow more secure than any other from commercial diversity libraries. The main focus at medicinal chemistry hand was fixed on the lead-like molecules to give more space for evolution of identified hits. At the same time, we were trying to focus on robust chemistry, which should not bring troubles with synthesis of scaffolds or their derivatisation. Our deep knowledge in synthetic chemistry already over 25 years’ experience allowed us to rate molecules by synthetic feasibility, which has been used as additional evaluation in the design of the largest Enamine Screening Library.
Key features
- No singletons, at least 3 compounds per cluster
- Immediate initial SAR, scaffold-secured
- Represents entire Enamine Screening Collection
- Lead-like compounds
- Hit confirmation & follow-up support
Along with all industry affiliated MedChem filters, sieve visual control of the core scaffolds (over 47k) have been applied. We believe that our effort will bring you a valuable hits. Some examples are represented below
Example of a cluster series
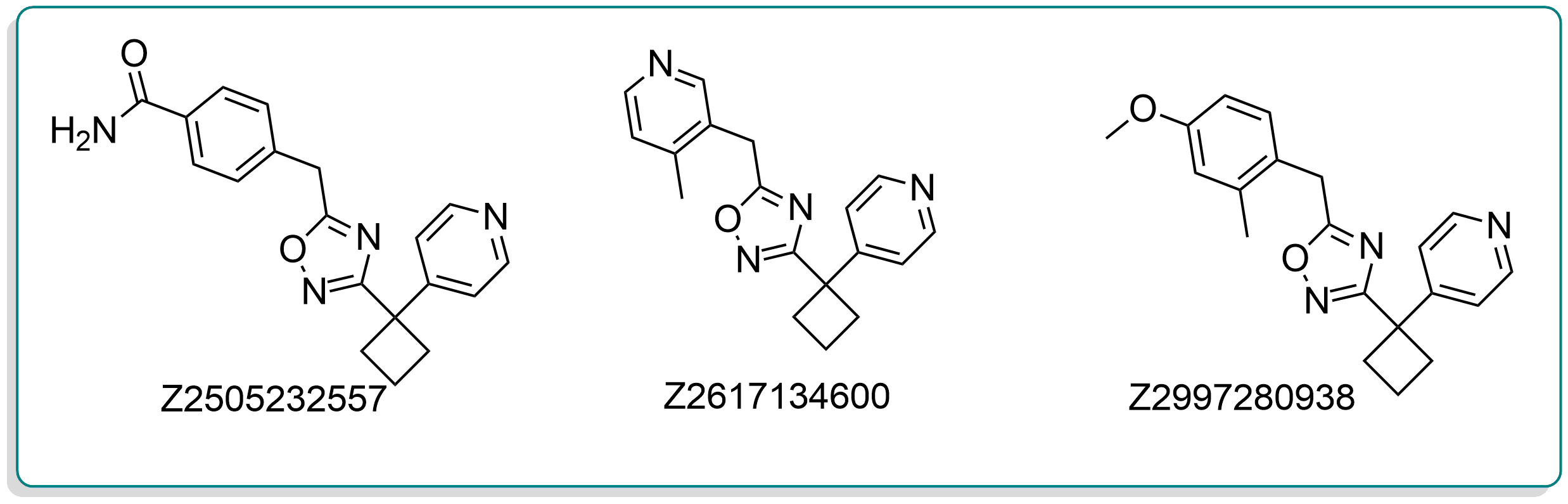
Cluster 458692
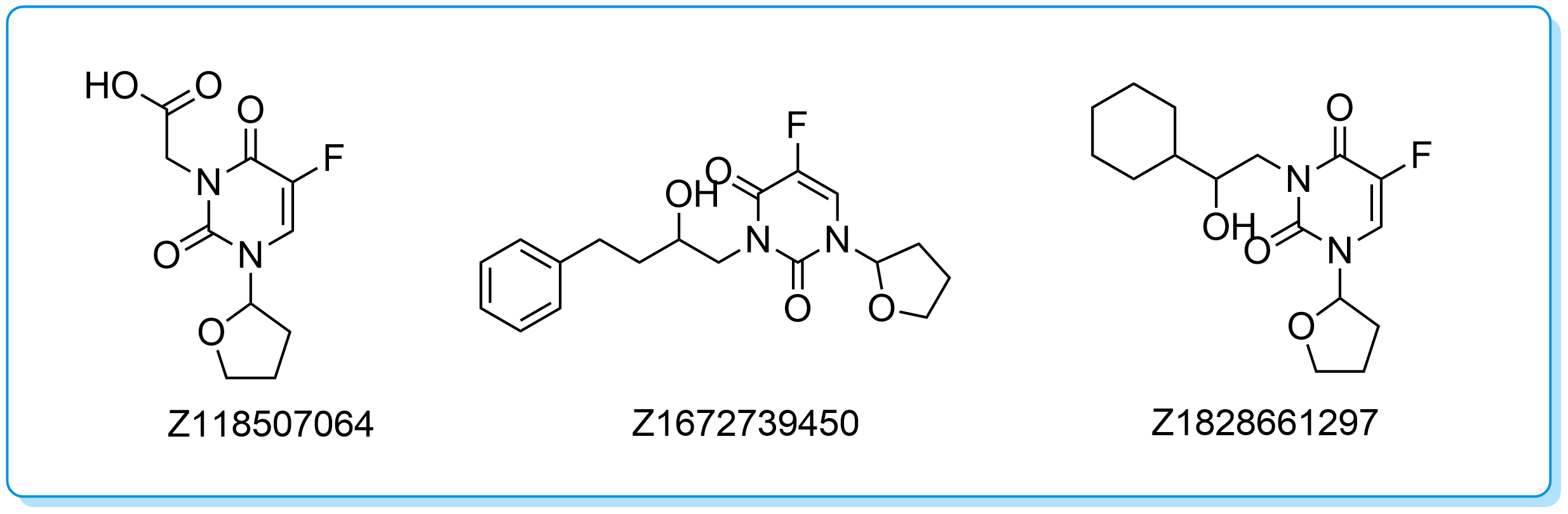
Cluster 103875
Representation of Hit Locator Library coverage of the entire Enamine stock Screening Collection
Top-quality diverse library of recently synthesized compounds
50 240 compounds
The success of high throughput screening in finding decent starting points for drug discovery heavily depends on the quality of the compound library. Compound screening libraries nowadays must bear an essential degree of novelty, possess contemporary lead-like properties and be built on “live chemistry” that can supply related chemical entities.
Enamine is constantly updating its Screening catalog with at least 225 000 newly synthesized compounds each year. These molecules are built on novel chemistry with use of the latest achievements in parallel chemistry and following industry trends in molecular properties and structural features. Every new library to be synthesized went through serial filters and is focused to bring novelty in a certain drug discovery area.
Discovery Diversity Sets (DDS) are aimed to bring latest chemistry to our customers within a smart clustering approach. Discovery Diversity Sets are based only on the lead-like compounds including representatives of all three main screening collections – HTS, Advanced, and Premium.
Typical Formats
Discovery Diversity Set is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
DDS-50-0-Z-10
Compounds
50 240
40 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
DDS-50-10-Y-10
Compounds
50 240
157 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
DDS-50-50-Y-10
Compounds
50 240
157 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
DDS-50-10-Y-10 screening library 50 240 cmpds, hit resupply, analogs from 4.3M stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
The hits from Discovery Diversity Sets are expected to be fast and easy to produce follow-up SAR libraries.
Download SD files
Key features
- Novel compounds
- No singletons, 3-5 compounds per cluster
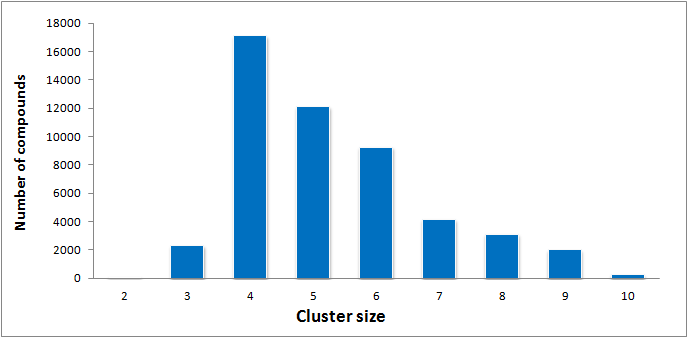
- 10 234 clusters in DDS-50
- No PAINS, only MedChem friendly compounds
- Express follow-up guaranteed
Distribution of clusters size (# of compounds with a cluster) in the library
Examples of molecules within a cluster
PPI-like scaffolds Cluster 4
GPCR-like structures Cluster 14
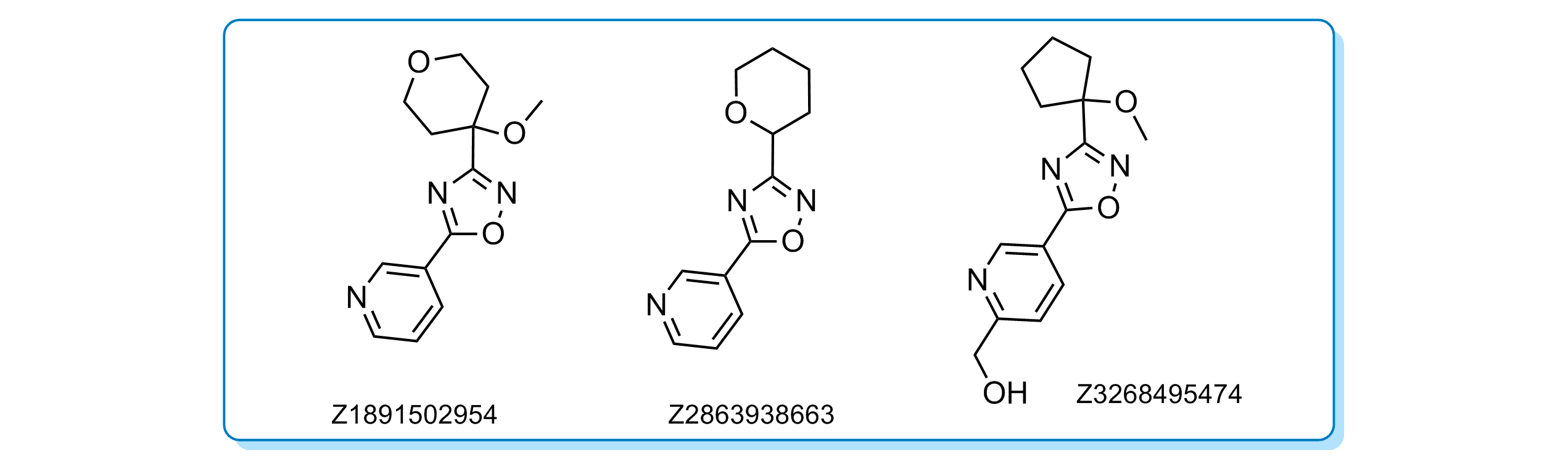
CNS-like scaffold Cluster 34
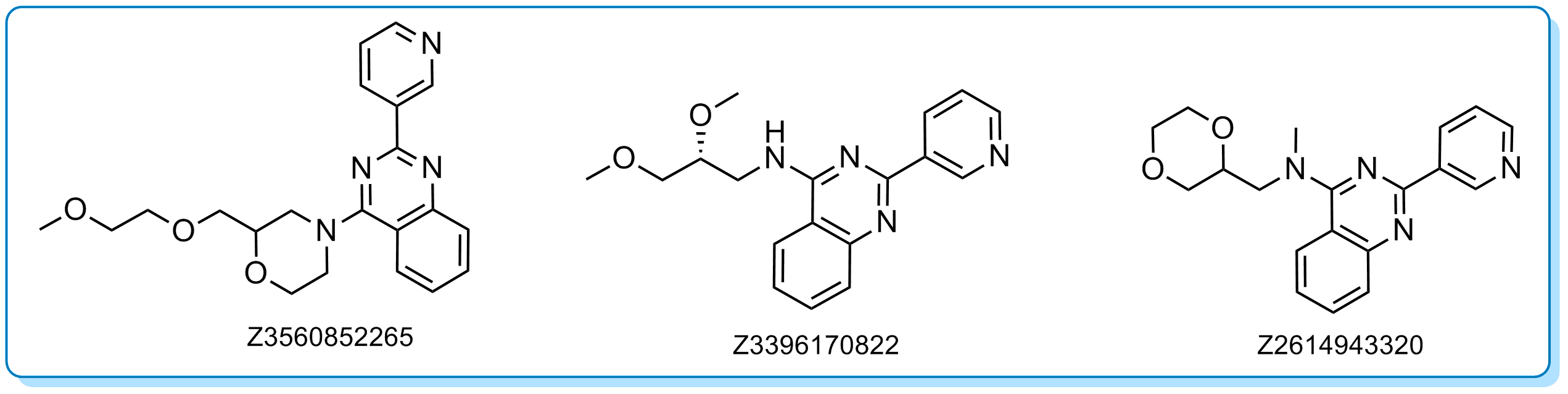
Kinase scaffold Cluster 11062