Designed for discovery of novel PPI inhibitors
40 640 compounds
Having a pivotal role in multiple cellular processes, protein-protein interactions (PPIs) are responsible for the effects linked to the blockage of the substrate binding site. There are numerous examples of drug candidates that were withdrawn from clinical trials due to the unexpected effects of direct antagonists. This fact outlines the importance of developing new potent protein-protein interactions inhibitors.
We have carefully selected 40 640 diverse compounds specifically targeting PPIs. All compounds are stored as dry substances and they can be acquired in diverse custom formats. Using our PPI Library for hit discovery you receive multiple benefits allowing you to save on time and costs in lead generation:
- Dry stock of over 4.7M compounds for hit resupply and hit expansion.
- Low-cost synthesis of analogues within only 3 weeks through our REAL Database technology
- Medicinal chemistry support enhanced with on-site broad ADME/T panel
You have also an option to screen the Library directly at Enamine. In this case, we will be happy to offer discount on library cost depending on the collaboration scope.
Typical Formats
Protein-Protein Interaction Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
PPI-40-0-Z-10
Compounds
40 640
32 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
PPI-40-10-Y-10
Compounds
40 640
127 plates
Amount
10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
PPI-40-50-Y-10
Compounds
40 640
127 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
PPI-40-10-Y-10 screening library 40 640 cmpds, hit resupply, analogs from 4.7M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library code: PML-8960
Version: 29 September 2023
8 960 compounds
sublibrary of PPI-40
Library design
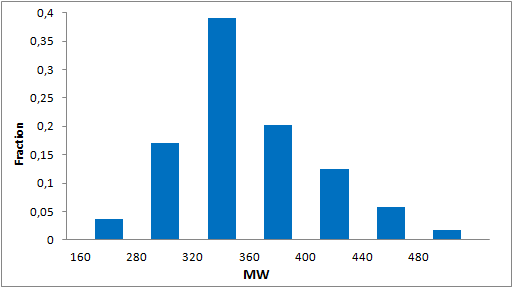
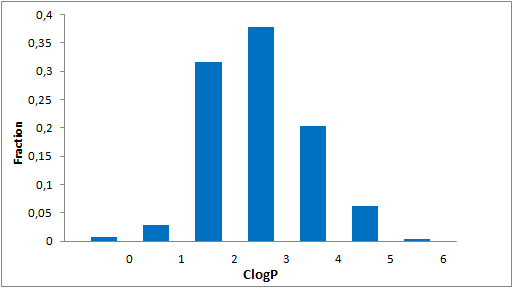
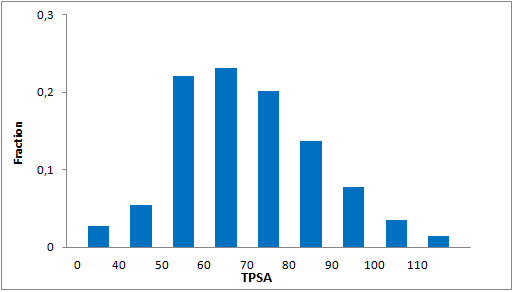
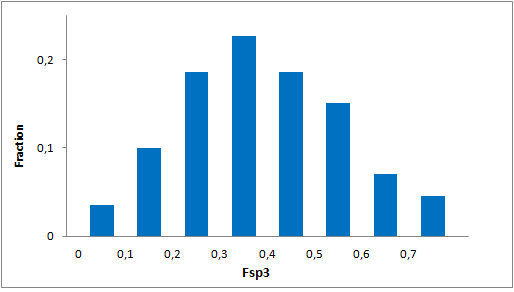
Systemic analysis of available structural data of numerous PPIs allowed us to develop dedicated approach to the library design. We have analyzed more than 20 different protein-protein complexes to highlight specific features of majority of potent inhibitors in this area. Several specific recognition patterns, like α-helix, β-sheet, PDZ-, PBD and bromodomains were used in the library design. As a result of Ligand- and Structure based in silico screening, we created the selection of compounds featuring:
- Specific recognition patterns, including hot spots analysis, key amino acids, secondary/tertiary structures, α-helices, ‘hot loops’ and specific protein domains affinity.
- Lead-like properties and sp3-rich core structural motifs. Compounds passed all including affiliated MedChem filters including PAINS.
- Latest chemistry and novel building blocks. Identified hits can be readily followed with synthesis of new analogs through REAL Database technology.
Examples of scaffolds populating PPI Library
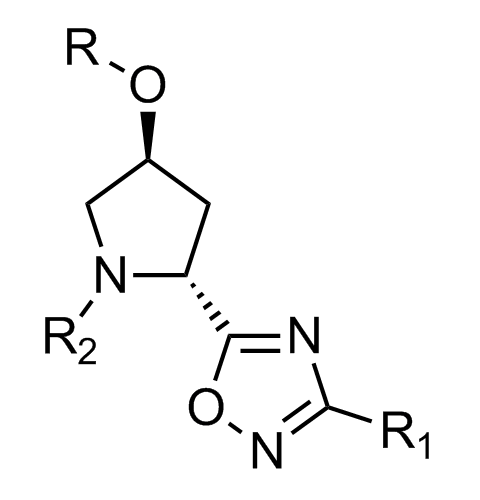
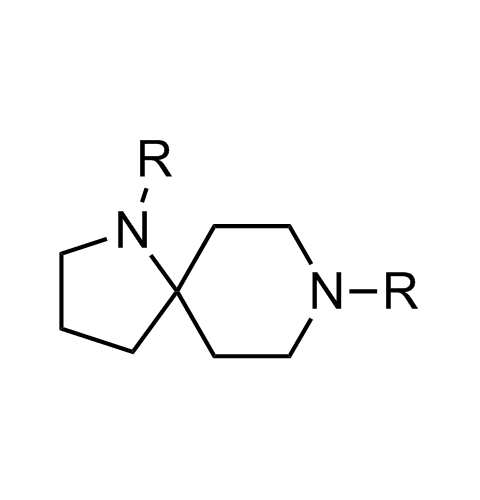
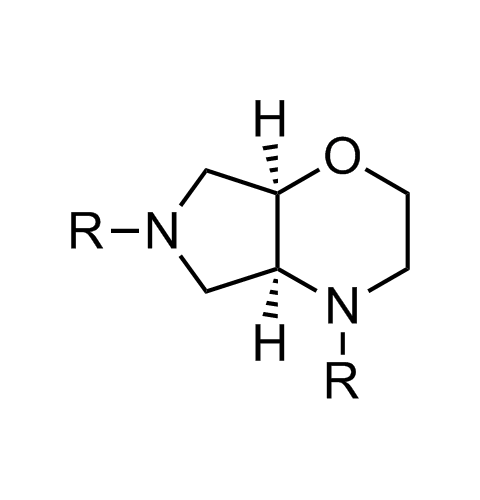
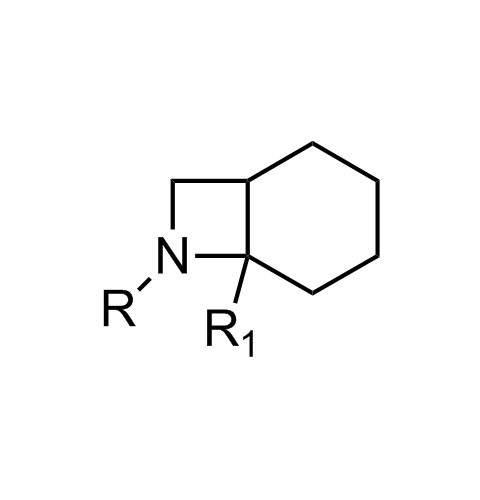
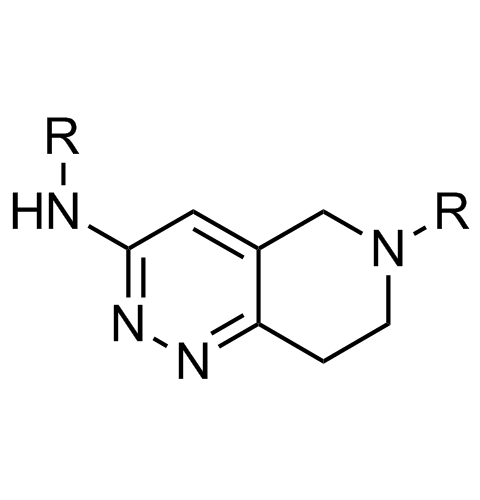
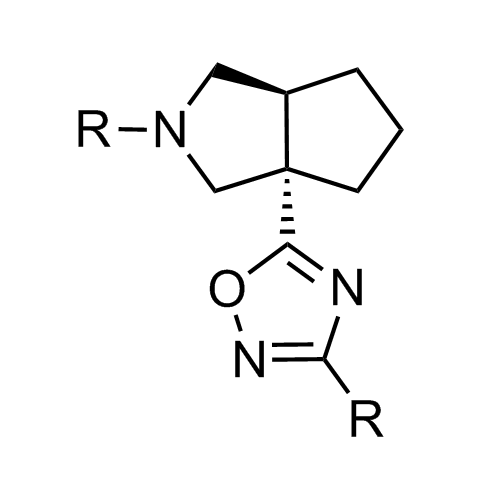
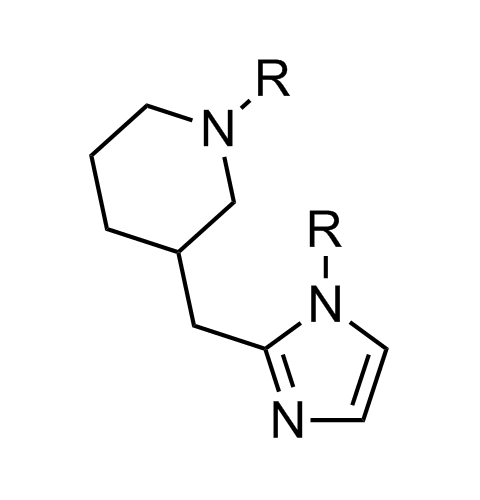
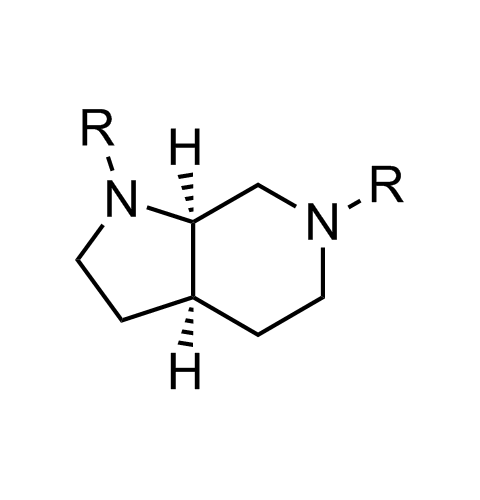
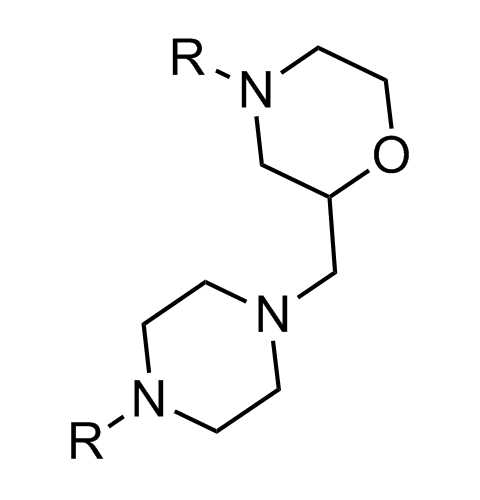
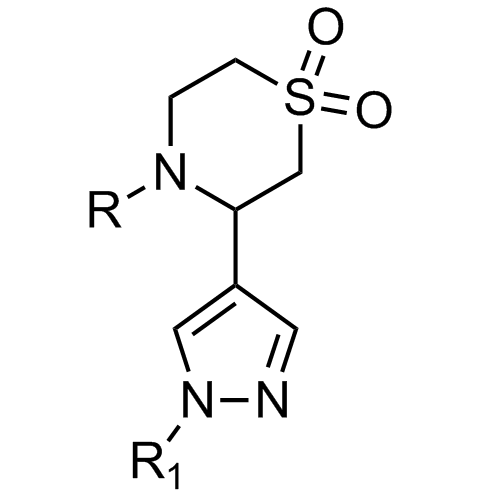
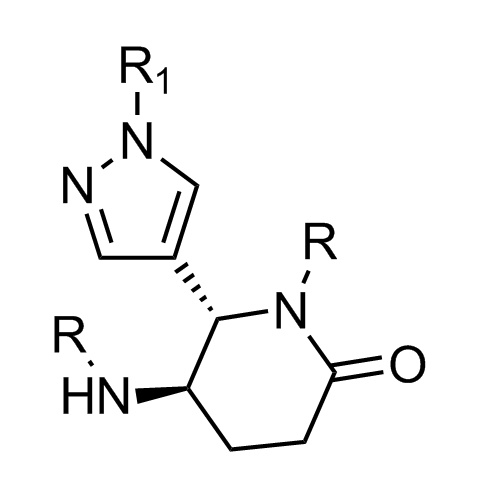
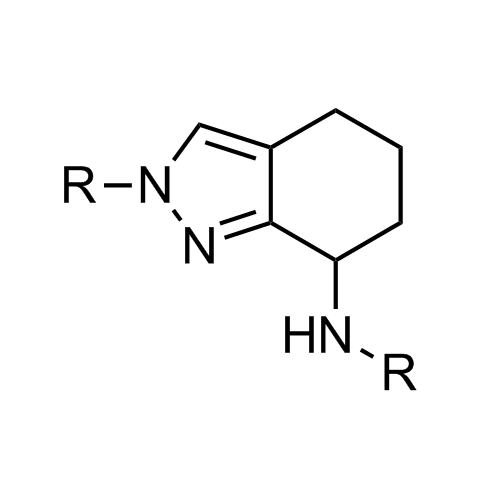
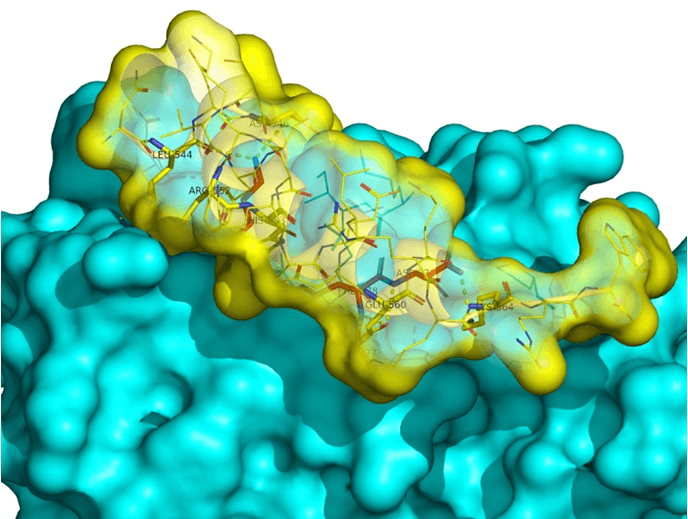
Importantly, our team of experienced computational, synthetic, medicinal chemists and biologists is ready to address your specific needs in tackling protein-protein interaction of your interest. Please, challenge us with your biological concepts, computational ideas and synthetic designs.