The effectiveness of the hit identification process depends on the screening method and screening library quality. To select current covalent and noncovalent fragment libraries with desirable properties, diversity analysis is typically utilised. ChemPass' UFrag platform is based upon pharmacophore, shape and vector analysis of vast ligand binding data in the protein database and it combines several key technologies that are necessary to revolutionise fragment library design.
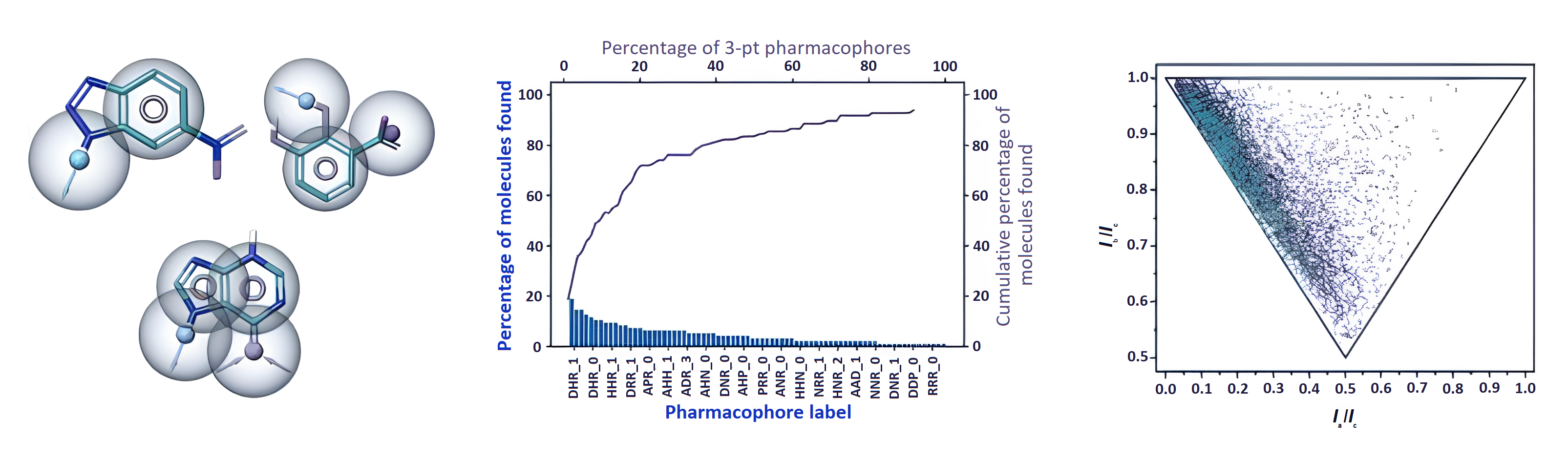
Conventional fragment libraries created through diversity analyses have been demonstrated to contain a significant number of redundant spacer and pharmacophore motifs, which leads to few to no hits against pockets of challenging targets and high hit rates of redundant rings for specific targets. The outcome is a significant danger of missed chances and a very varied experimental hit rate.
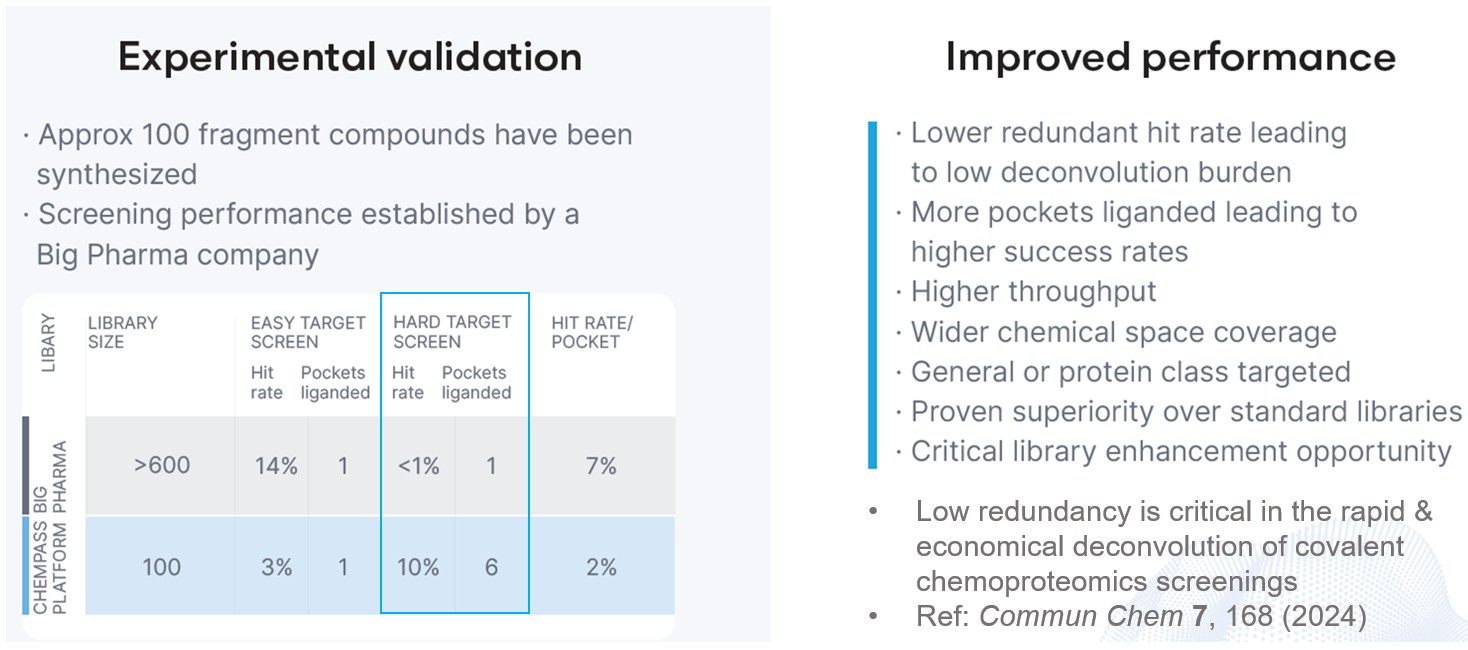
The platform's inherent customisation features allow the library design to target particular protein classes while minimising both the size of the library and the expense of synthetic materials. In order to confirm the notion, a test library was screened against a validated and hard target. The results showed that the test library produced a much higher hit rate and binding site coverage against the hard target than a standard fragment library.
Why UFrag™ library from Enamine & ChemPass?
- Higher success against tough targets.
- Less redundancy for easy targets.
- Superior pharmacophore analysis – better coverage, higher hit rates, fewer redundant compounds.
- Optimized fragment growth – improved hit rates and optimizability.
- More efficient vector analysis – fewer compounds, faster drug discovery.
- Short synthesis timelines, low costs, and high laboratory success rates.
UFrag™ Libraries
As an additional proof-of-concept, together with ChemPass we have developed two general non-covalent fragment libraries: the UFrag™ General Library and the UFrag™ Essential Library. ChemPass’ Universal Fragment Library (UFL) Design Platform, combined with additional MedChem filtering, was applied to over 300 000 Enamine fragments. Almost all fragments included in the final version of the libraries are available in quantities of 50 mg or more. The stereochemical nature of all library members is either a single enantiomer, a racemate, or a compound without stereoisomers—there are no diastereomeric mixtures.
The UFrag™ Essential Library, containing 372 compounds, is a minimalistic set of pharmacophore-optimized fragments specifically designed for maximal UFL cluster space coverage. Its small size is expected to facilitate efficient screening.
The UFrag™ General Library, comprising 2 301 compounds, is an extended version of the smaller set. It is designed to broadly cover fragment space and provide greater shape and scaffold diversity.
Both libraries can be offered as 100 mM DMSO solutions or as dry powders.
Typical Formats
Catalog No.
UFrag™ General Library
UFLg-25-Y-100
Compounds
2 301
8 plates
Amount
25 µL of 100 mM DMSO stock solutions
Plates and formats
384 well, Echo Qualified LDV microplates 001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
UFrag™ Essential Library
UFLe-50-Y-100
Compounds
372
2 plates
Amount
50 µL of 100 mM DMSO stock solutions
Plates and formats
384 well, Greiner Bio-One microplates 781280, first and last two columns empty, 320 compounds per plate
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400) or send your preferred labware. Compounds pooling can be provided upon request.

