Anti-Cancer Agents in Medicinal Chemistry , 2007, 7 (2), 171-188
DOI: 10.2174/187152007780058704
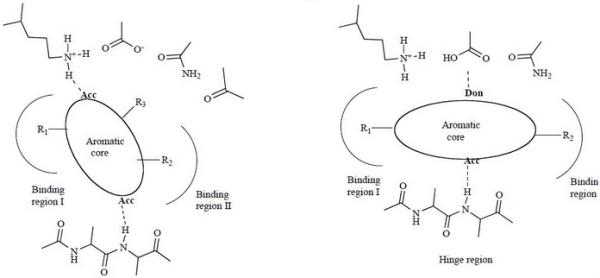
Protein kinases are among the most exploited targets in modern drug discovery due to key roles these enzymes play in human diseases including cancer. The in silico approach, an important part of rational design of protein kinase inhibitors, is founded on vast information about 3D structures of these enzymes. This review summarizes general structural features of the kinase inhibitors and the studies applied toward a large scale chemical database for virtual screening. Analyzed are the ways of validating the modern docking tools and their combinations with different scoring functions. In particular, we discuss the kinase flexibility as a reason for failures of the docking procedure. Finally, evidence is provided for the main patterns of kinase-inhibitor interactions and creation of the hinge-region-directed 2D filters.
Dubinina G.; Chupryna O.; Platonov M.; Borisko P.; Ostrovska G.; Tolmachov A.; Shtil A.
Anti-Cancer Agents in Medicinal Chemistry 2007, 7 (2), 171-188
DOI: 10.2174/187152007780058704
