A sub-library of Enamine’s GPCR Library designed for discovery of novel lipid GPCR ligands
6 400 compounds
Enamine’s Lipid GPCR Library has been designed in collaboration with NQuiX. Over 6 400 compounds have been selected from off-the-shelf collection or specifically synthesized for this project from Enamine REAL screening Collection. They were designed to target the family of eight endothelial differentiation gene (EDG) receptors (S1P1–5 and LPA1–3). The compounds may also be appropriate for screening at GPR3, GPR6 and GPR12 orphan receptors given some TM bundle binding site similarity and/or GPR23, GPR92 and P2Y5 given their more recent classification as additional, albeit distinct, LPA receptors.
Typical Formats
Lipid GPCR Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
LGR-6-0-Z-10
Compounds
6 400
5 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
LGR-6-10-Y-10
Compounds
6 400
20 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
LGR-6-50-Y-10
Compounds
6 400
20 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, first and last two columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
LGR-6-10-Y-10 screening library 6 400 cmpds, hit resupply, analogs from 4.6M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library design
The compounds have been selected using a combination of ligand-based methods using chemical fingerprints, 2D pharmacophores and 3D shape/feature matches (Figure 1). The training active compound set included 1 393 compounds (870 acidic and 523 non-acidic), collected across 73 papers (Figure 2). They represent a combination of compounds for expanding SAR around known chemotypes and scaffold hops seeking novelty. The current set of compounds covers the non-acidic classes of ligand, with acidic compounds being designed via a different procedure and added to the library separately.
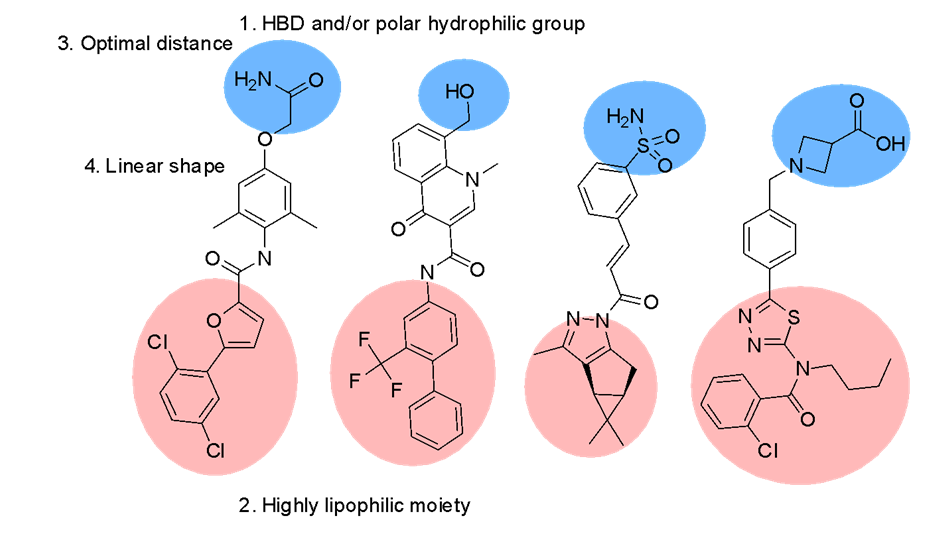
Fig. 1.Dedicated design of the compounds included in the Library (general scheme).
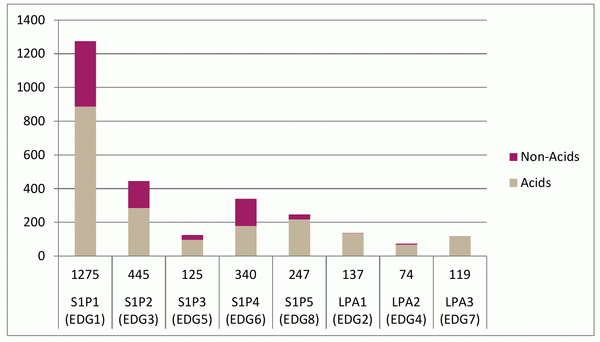
Fig. 2. Distribution of the compounds in the training set.