The interest of pharmaceutical companies in DNA-encoded chemical libraries (DEL) technology has been growing over the years, with numerous organizations now having their own screening programs using DELs, or outsourcing capabilities from specialized DEL providers.
A bit of history
The concept of DNA-encoded libraries (DEL) appeared in the early 1990th when scientists from Scripps Research Institute published a paper on “Encoded combinatorial chemistry”. The main principle of DEL technology is to encode each compound in the library with a unique DNA barcode. Thereby a library of billion compounds can be screened as a mix in a single test tube and the active compounds are identified by amplification and sequencing of the DNA barcodes.
At the initial stage, companies pioneering in DEL development were faced with a lack of relevant methods for DNA sequencing. Scientists from Praecis Pharmaceuticals then founded a company which managed to sequence DEL libraries. Praecis and Nuevolution were the first to have taken a chance on DEL in the 2000s. In 2005 and subsequent years the second generation sequencing has emerged making DEL a much more attractive and economically feasible approach in drug discovery.
An overview of DEL technology
DEL technology comes from the merge of DNA encoding and combinatorial chemistry. The most straightforward method to build DEL is the “split and pool” approach. At the first step of the synthesis, chemical building blocks (BBs) are tagged with DNA barcodes. In the second step, they are mixed together and then split into different portions. Next, a new set of building blocks is added to the chemical reaction, and then, corresponding DNA barcodes are attached and linked with previous DNA molecules. Thus, DNA encodes each step of chemical synthesis for each compound.
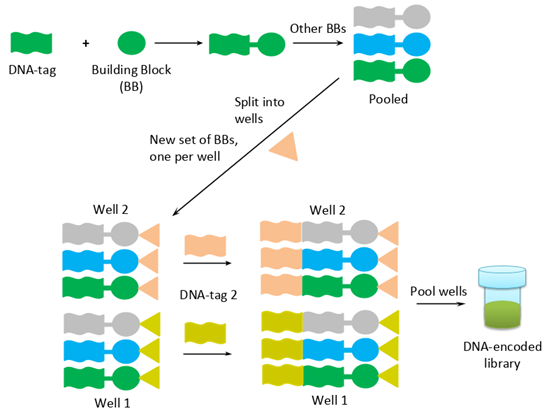
* Reproduced from C&EN
One of the more sophisticated methods to build libraries is to use DNA code to determine the sequence of chemical reactions. Templates are prepared before synthesis and contain regions complementary to barcodes of BBs. When barcodes couple to the template, the relative sterical arrangement of building blocks facilitates a chemical reaction between them. This method is particularly useful when assembling libraries of macrocycles and peptides. The alteration of the approach is used in Vipergen’s YoctoReactor.
Size and costs of DELs
“Split and pool” method enables building astronomically large libraries containing up to 10^10 compounds. For example, HitGen’s libraries contain more than 1 trillion molecules, X-chem provides a collection of over 18 libraries covering over 7.5 billion compounds. for screening, and Nuevolution assembled a collection of 40 trillions compounds (in 2019, Neuvolution was acquired by Amgen for $167 million). To compare, the size of a common high throughput screening (HTS) library is restricted to several million compounds due to the prohibitive cost of synthesizing and managing larger collections. Overall, the cost of creating and screening an HTS library of 1 million compounds would cost millions of dollars (approximately $1,100 per compound), while screening a DEL library of 800 thousand compounds would cost on the order of $150.000.
Hit discovery using DELs
Screening of the DELs is conducted in single test tubes, where the target protein is incubated with a library. The target proteins can be anchored on a solid support, commonly magnetic beads. After thorough washing, only high-affinity ligands remain associated with a target. After DNA sequencing and decoding of the structure of active compounds, the last should be resynthesized to repeat the affinity binding assay. A lot of different selection strategies can be applied, even phenotypic screening is compatible with DELs.
DEL is suitable for the synthesis and screening of small molecules, macrocyclic compounds, and peptides. Scientists are harnessing chemical properties of DNA, especially the principle of complementarity, for developing new strategies for library design.
Convergence with artificial intelligence
Since DEL technology offers access to essentially the largest chemical space available on the market, this big data technology is a natural fit for big data analytics and modeling technologies offered by the field of artificial intelligence -- and some companies are chasing this opportunity.
A notable deal took place in 2020, when Insitro, a leading player in the application of machine learning for drug discovery, founded by Daphne Koller, acquired Haystack Sciences. Haystack’s machine learning-based platform combined multiple elements of their DEL technology, including the capability to synthetize broad, diverse, small molecule collections, the ability to execute iterative follow-up, and a proprietary semi-quantitative screening technology, called nDexer™, that generates higher resolution datasets than possible through conventional ‘panning’ approaches.
In 2020, Google Research published results of their collaborative effort with X-Chem, one of the leading players in the DEL technology space, in an article “Machine learning on DNA-encoded libraries: A new paradigm for hit-finding”, where they demonstrate an effective new method for finding biologically active molecules using a combination of physical screening with DNA-encoded small molecule libraries and virtual screening using a graph convolutional neural network (GCNN). This research has led to the creation of the Chemome initiative, a cooperative project between our Accelerated Science team and ZebiAI, a platform that applies massive experimental DNA encoded library data sets to power machine learning for drug discovery.
In its turn, ZebiAI was acquired in 2021 by another notable developer of artificial intelligence-powered drug discovery platform, a clinical stage biotech Relay Therapeutics (NASDAQ: RLAY), where Relay paid $85 million up-front. This acquisition allowed Relay to incorporate ZebiAI’s machine-learning-based DEL technology into their protein targeting platform Dynamo.
In October 2021, X-Chem acquired Glamorous AI, a developer of a modular multifaceted artificial intelligence solution for drug discovery RosalindAI, including capabilities of data engineering and featurization, predictive analytics, high-performance computing, and de-novo drug design.
Notably, half a year later, Enko, the crop health company, announced its acquisition of proprietary DNA-encoded libraries and technology enablement from X-Chem. With that deal, Enko announced to be the first company to internalize this technology for agricultural applications. DNA-encoded libraries are a proven tool to kickstart pharmaceutical drug discovery projects because of the efficiency of screening large, diverse chemical spaces. Enko pioneered applying this approach to tackle agriculture challenges. The company said this acquisition is a significant advancement for Enko's ENKOMPASS™ platform, which combines DNA-encoded library screening with machine learning and structure-based design to find novel chemistry and new modes of action to control crop pests and diseases.
HitGen is a Shanghai Stock Exchange listed company, one of the leaders in the DEL technology market. In November 2021, HitGen and Cambridge Molecular announced a strategic partnership, bringing together HitGen’s DNA-encoded libraries and DeepDELve 2 – Cambridge Molecular’s highly optimized DEL-specific deep learning system. DeepDELve 2 is fully integrated with Enamine REAL space, Mcule, Chemspace, and eleven other compound catalog providers, and can be linked on demand to commercial or internal catalogs of choice. As such, potential ligands are often delivered to a partner’s lab in as little as a few days.
Another notable project to implement machine learning on large DNA-encoded libraries, “DEL-ML-CS”, was implemented by ChemSpace, a leading chemical provider of small molecules for pharmaceutical research. The team at ChemSpace decided to implement the idea, proposed by Kevin McCloskey et al. in a 2020 paper “Machine Learning on DNA-Encoded Libraries: A New Paradigm for Hit Finding” to combine machine learning and DNA-encoded libraries (DEL) screens. Using a DEL screen, it is possible to access vast chemical space in a single experiment. This helps to get substantial amounts of data faster and with less resources required. This data can be further used to build a relevant machine learning model and then apply it on the ultra-large chemical space of small molecules provided by ChemSpace -- allowing for a high probability to identify potent hits from the first screen.
Within the scope of this project, ChemSpace provides access to OpenDEL, a HitGen’s new encoded library of over 145 million drug-like small molecules that can be screened using only 2 mg of target protein, at costs comparable to HTS on much smaller decks. The OpenDEL library is based on 70 different templates and is designed for high diversity within drug-like space.
The above-mentioned collaborations illustrate a growing interest in the application of artificial intelligence to DNA-encoded chemistry -- for drug discovery and agricultural applications.
DEL-compatible chemistry
A very important aspect of DEL technology is the underlying chemistry and certain limitations imposed by the very properties of DNA.
DNA is a water-soluble molecule, thereby traditional organic reactions should be translated to corresponding conditions. In the beginning, only a few reactions were available in the toolbox of chemists, but their number is extended, harnessing various synthesis approaches. For example, one of the recent ideas was to employ photoredox catalysis for DNA‐encoded chemistry.
At Enamine, we have a diverse range of DNA-compatible scaffolds, an indispensable supporting tool to enable DEL projects of any size and complexity. The current virtual database includes 175 thousand of bifunctional and 7.5 thousand of trifunctional building blocks. All molecules in this space are feasible for synthesis with high predicted success rate, since their enumeration is based on previously acquired validated experimental data. Additionally, custom synthesis of specific core-structures for the Customer’s needs are also possible and could be performed on both FFS and FTE bases. Our team of experienced Synthetic Organic Chemists can deliver new desired molecules at competitive prices in a short lead time.
The on-DNA synthesis of functionally diverse pyridazines based on inverse-electron-demand Diels-Alder (IEDDA) reactions of 1,2,4,5-tetrazines has been shown recently. Herein we propose tetrazine-derived compound library designed for DNA ligation and DNA-compatible IEDDA reactions. See also: "Structurally optimized tetrazines for rapid biological labeling"
