Designed to promote the discovery of new-generation medicines
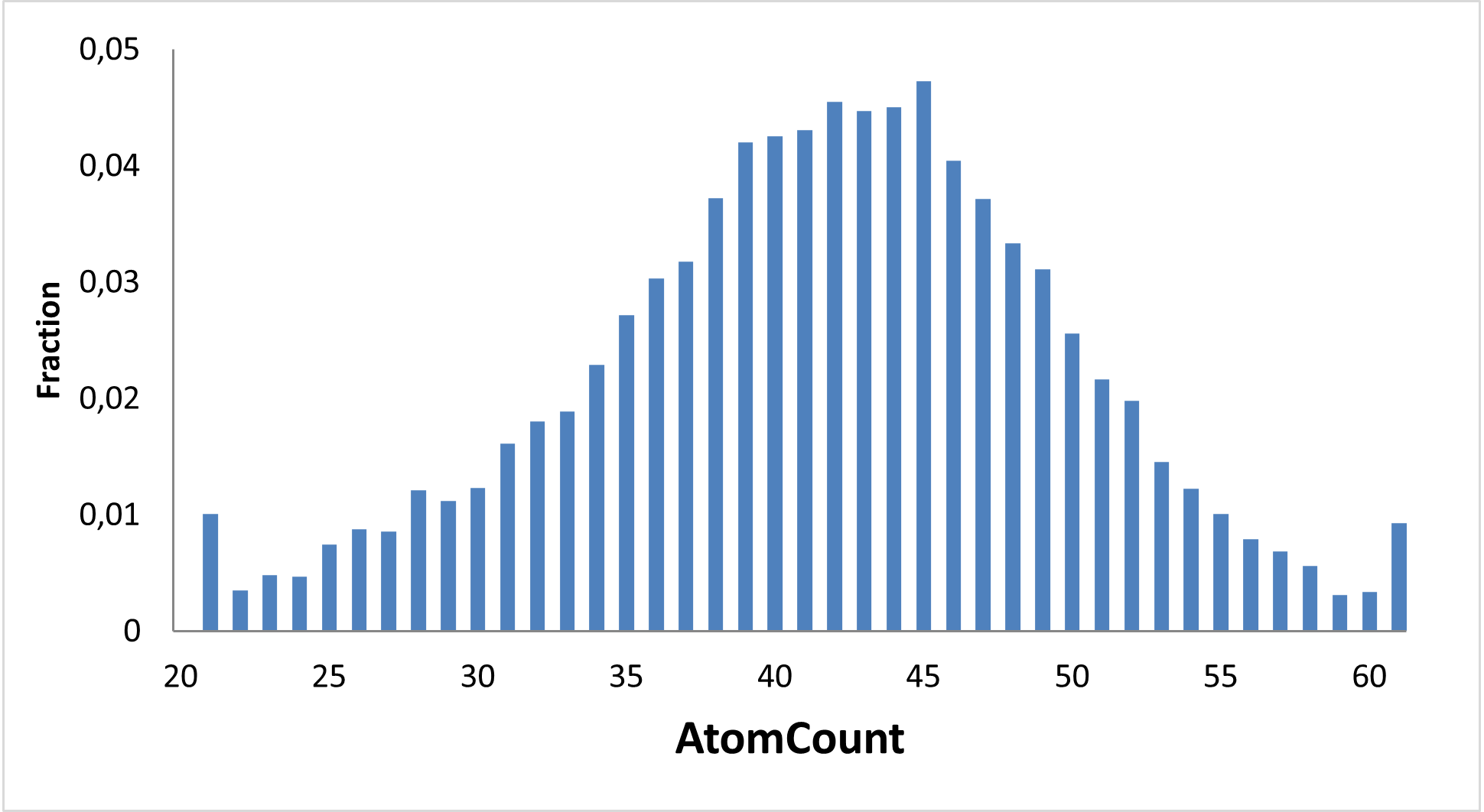
28 000 compounds
RNA Splicing Library 10 400 compounds
RNA Riboswitches 14 800 compounds
RNA G-quadruplexes 2 800 compounds
RNA-focused drug discovery has dramatically changed over the last decade and has become the most exciting frontier in developing new-generation pharmaceuticals for better healthcare. This new stage of RNA investigation, also called “RNAissance” owes to recent advances in genetic engineering and genome sequencing. This gives us high anticipation for discovering efficient medications against difficult-to-treat and incurable diseases, along with new antibacterials and antivirals.
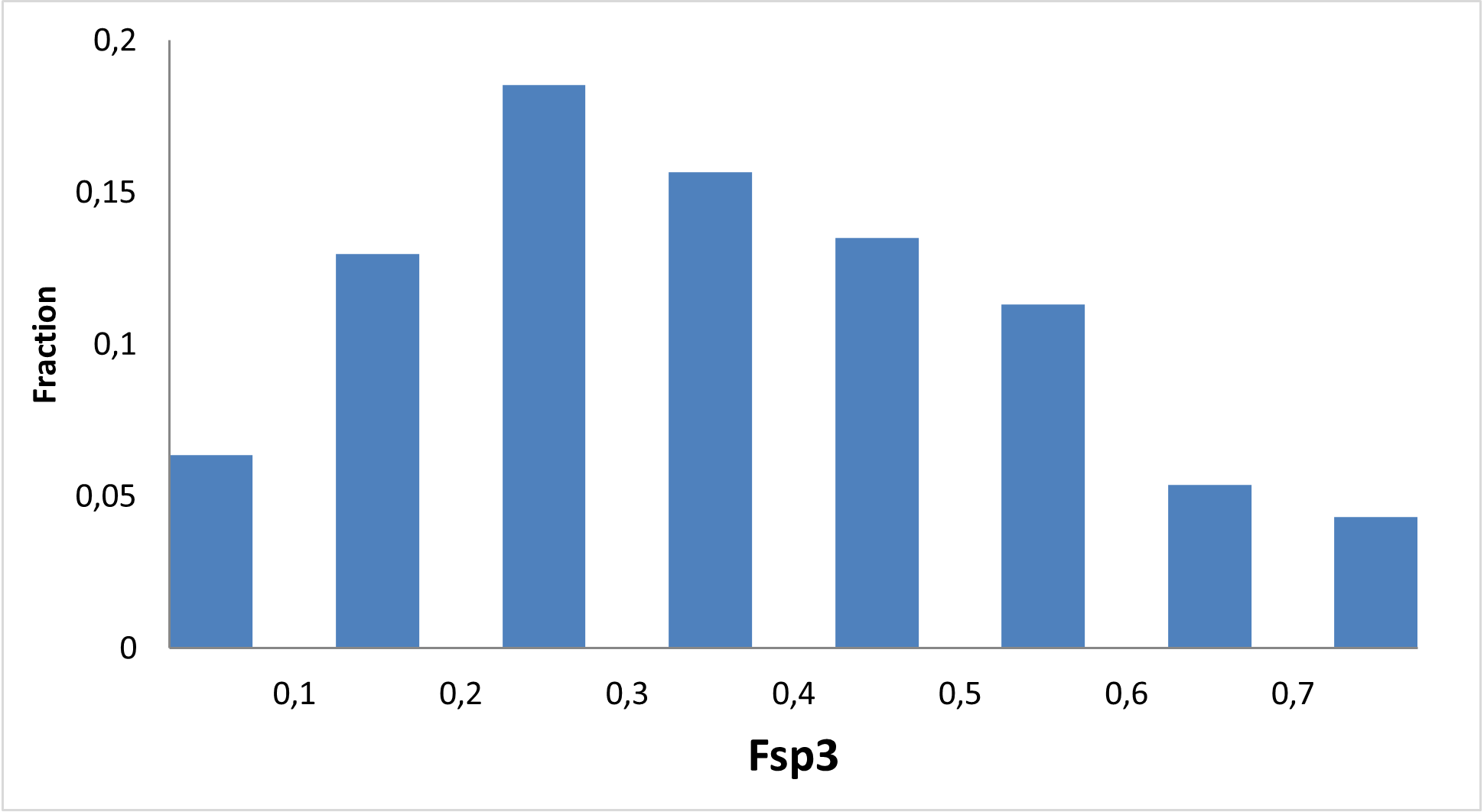
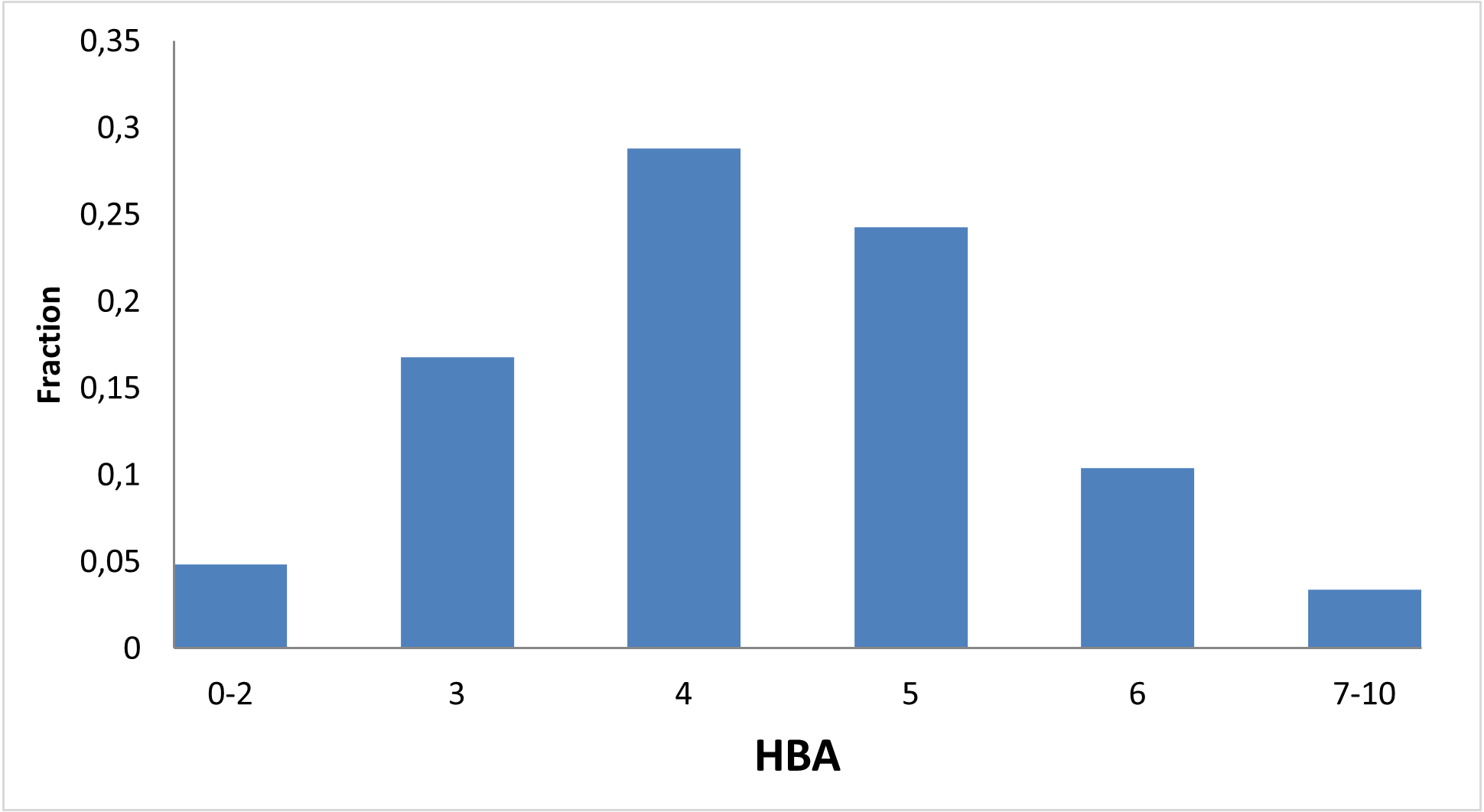
RNA is a versatile biomolecule capable of transferring information, adopting distinct three-dimensional shapes, and responding to ambient conditions. These biological structures can carry out a wide range of functions, from translating genetic information into the molecular machines and structures of the cell to regulating the activity of genes during development, cellular differentiation, changing environments, and more.
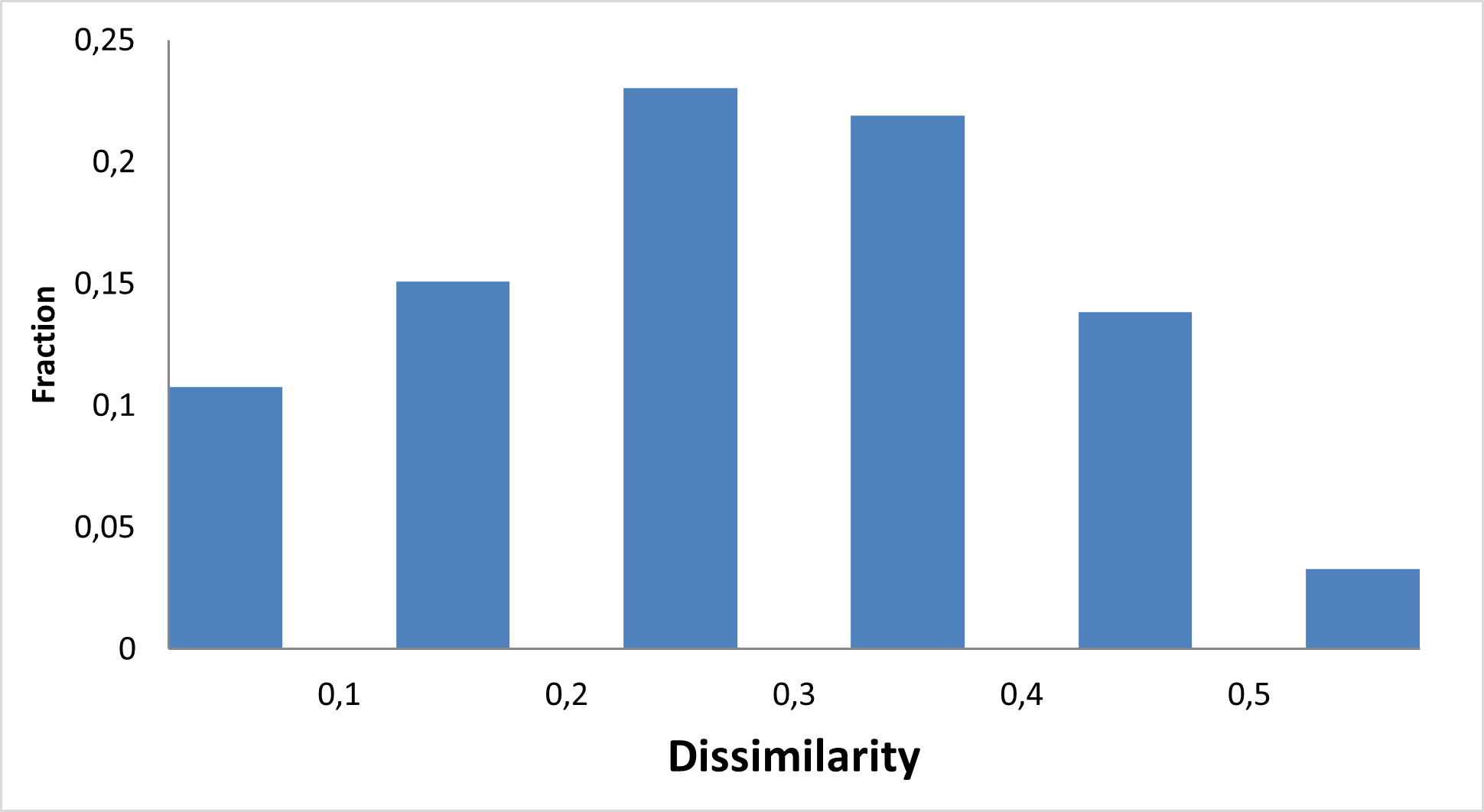
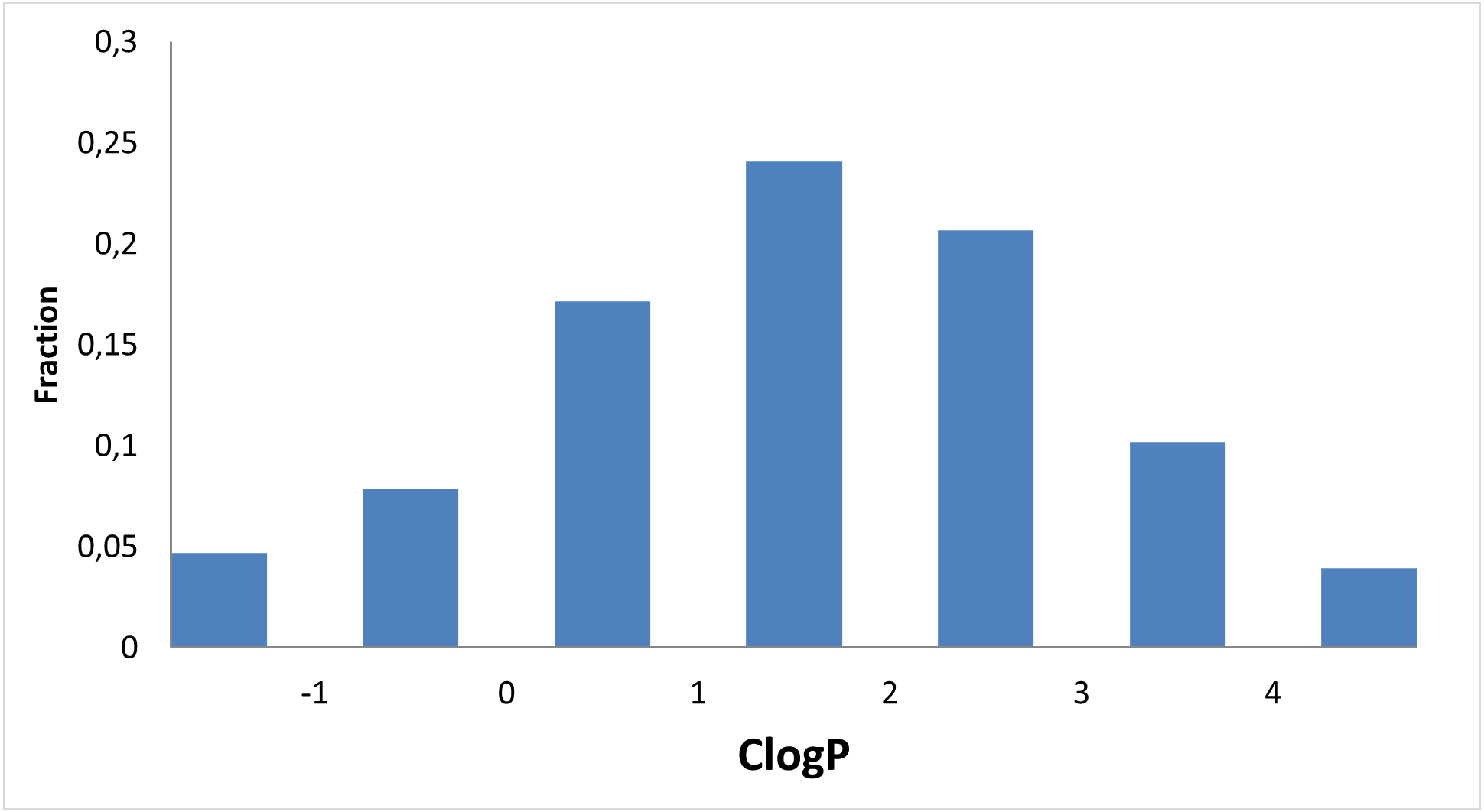
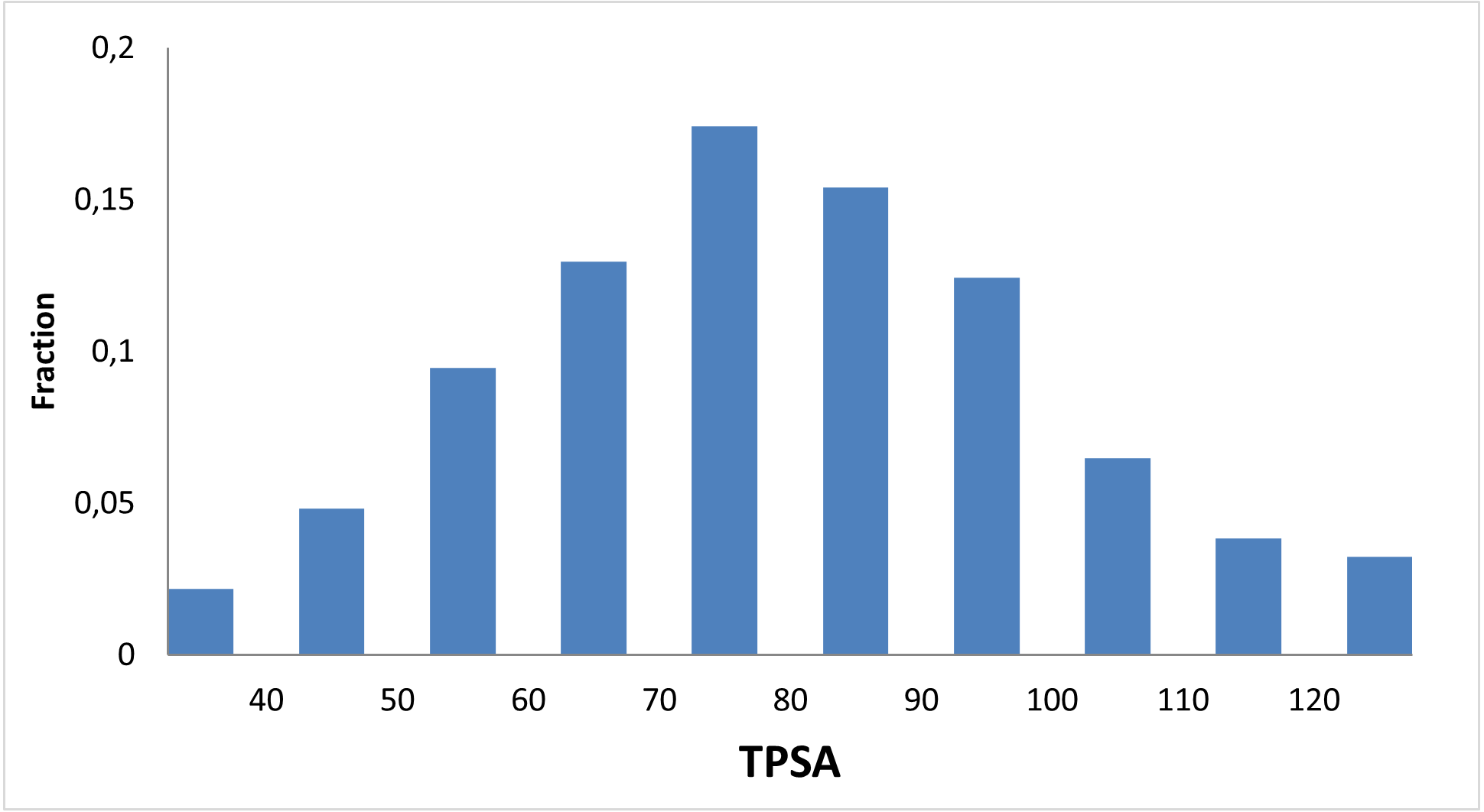
We focused on investigating three main types of interactions of small molecules with RNA secondary structures and 3D fold of tRNAs, and mRNAs (avoiding direct targeting of RNA helix and intercalation) with virtual screening of available structures of G-quadruplex and Riboswitches.
The library has been plated for the most convenient and quick access. Using our RNA library you receive multiple benefits, allowing you to save on time and costs in hit exploration and optimization:
- Hit confirmation support: Analogs search and hit samples resupply from dry stock with strict QC
- Straightforward & affordable synthesis of hit follow-up libraries by parallel chemistry
- Medicinal chemistry support enhanced with on-site broad ADME/T panel
Typical Formats
RNA Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
RNA-28-5-Y-10
Compounds
28 000
88 plates
Amount
5 µL of 10 mM DMSO solutions
Plates and formats
384-well acoustic LDV microplates, 320 compounds per plate, 1, 2 & 23, 24 columns empty
Price
Catalog No.
RNAS-10400-10-Y-10
Compounds
10 400
33 plates
Amount
10 µL of 10mM DMSO solutions
Plates and formats
384-well, echo qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
RNAR-14800-20-Y-10
Compounds
14 800
47 plates
Amount
20 µL of 10 mM DMSO solutions
Plates and formats
384-well plates Greiner #781280, 320 compounds per plate, first two and last two columns empty
Price
Catalog No.
RNAG4-2800-50-X-10
Compounds
2 800
35 plates
Amount
50 µL of 10mM DMSO solutions
Plates and formats
96-well plates, Greiner #650160, 80 compounds per plate, 1 & 12 columns empty
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD files
Library design
G-quadruplexes are high-order, four-stranded structures formed by repeated guanine tracts in the genome. Despite the significant prevalence of DNA G-quadruplexes over RNA, RNA quadruplexes remain crucial in various biological processes such as epigenetic regulation, telomere maintenance, mitochondrial RNA expression, and many others. We analyzed all reported RNA G-quadruplexes (G4s) structures and created 6 docking models based on RNA aptamers and modeling of h-MYC G4s.
Riboswitches are bacterial regulatory segments of mRNA and consist of a metabolite-binding aptamer and an expression platform. They are essential for different processes: expression of viral genes, gene-regulating function, production control of the proteins encoded by the mRNA, catalyze reactions, bind to other RNA or regulatory macromolecules, and many more. Given their absence in mammalian cells, riboswitches are very attractive targets for the discovery of new antibacterials. Over 20 docking models have been created and used for in silico screening of the following riboswitches: 2-deoxyguanosine, c-di-AMP, c-di-GMP, class I PreQ1, FMN, glmS ribozyme with G40A mutation, Glutamine II, Glycine, Guanine, L-glutamine, Lysine, metY SAM V, preQ1 Class II, PreQ1, PRPP, s-adenosylhomocysteine, SMK box (SAM-III), THF, thi-Box, thi-M, ZTP riboswitch.
RNA splicing regulation is probably the most attractive in eukaryotic drug discovery. Modulation of RNA splicing would be one of the most effective ways for cancer treatment and genetic disorders. We focused on the targeting of regulatory and precursor mRNAs and kinases, which activity required for RNA splicing: serine and arginine-rich splicing factor protein kinases SRPKs and CDC2-like kinases CLKs.
The structure-based approach (molecular docking) has been used as a main technique for library construction. All available PDB structures of RNA G4s, riboswitches, mRNAs and SRPK and CLK kinase swere analyzed. Unique "protein-ligand" complexes were selected for further analysis and docking model constructions. The following PDB structures were used for G4s: 6C63, 6e81, 6E81, 6E8S, 7oa3, 8EYV; and Riboswitches: 2hom, 2miy, 3b4b, 3d0u, 3e5e, 3irw, 3npn, 3p49, 3q50, 3ski, 4lvy, 4nya, 4nyb, 4nyd, 4qk8, 4rzd, 5c45, 5ddq, 6ck4, 6fz0, 6p2h, 6qn3, 6uc8, 6uc9, 6wzs.
Pharmacophore models are represented below using the following color scheme: red spheres - H-bond acceptors, blue – Hb donors, orange – aromatic groups, and grey spheres represent any other atom types, purple – mixed acceptor/donor groups. RNA nucleobases are highlighted in green, the native is yellow, and an example of a docked ligand is shown in grey.
Examples of molecular docking simulation, and used pharmacophore models, G-quadruplex
Example 1 (PDB ID 5bjp, RNA aptamer)
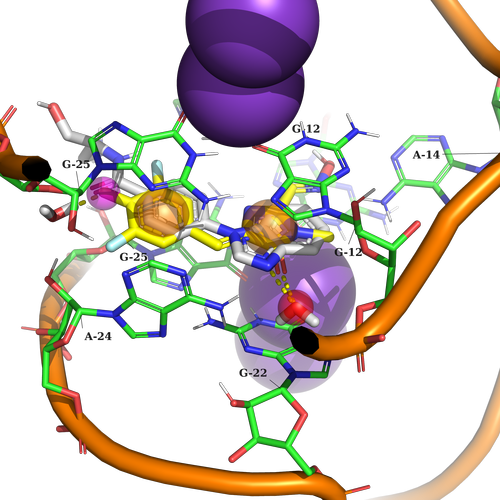
Example 1. The potential ligand should contain two aromatic features to create stacking interaction with G25, A24, G12, and G22. Additionally, it should be capable of forming hydrogen bond interactions with two HOH (water) molecules.
Example 2 (PDB ID 6c63)
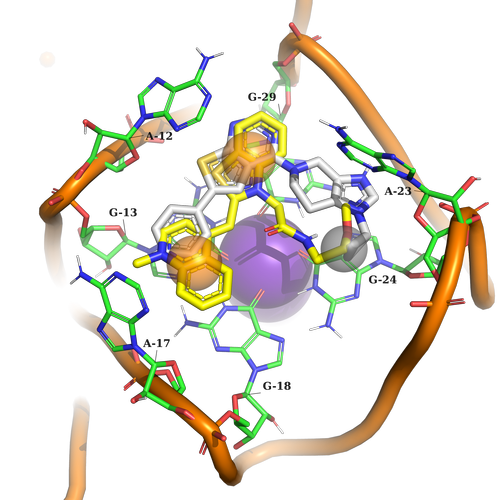
Example 2. The ligand must contain two aromatic parts for stacking interaction with G13, G18, A12, and G29. At another part of the pocket, it should just fill in the sub-pocket formed by A23 and G24.
Examples of molecular docking simulation, and used pharmacophore models, riboswitches
Example 1 (PDB ID 3e5e, SAM Riboswitch)
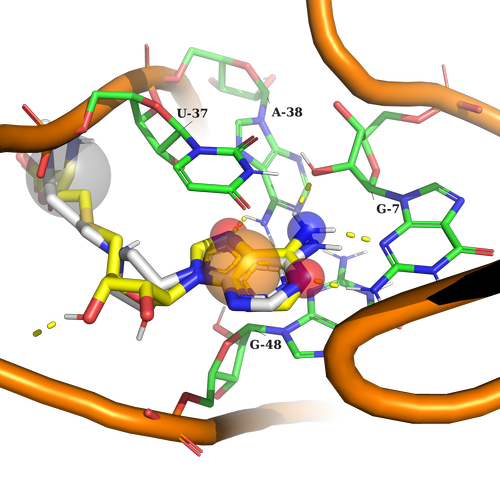
Example 1. According to the used pharmacophore model ligand should contain an aromatic part to interact by stacking interactions with U37, and G48, Any atom part just to fill in the sub-pocket behind U37, and 3 donors of h-bond interaction (two acceptors, and one donor) to interact with G7, A38. It is important to note here, that the ligand should contain at least 2 of 3 h-bond interaction features used in the pharmacophore model.
Example 2 (PDB ID 3d0u, Lysine Riboswitch)
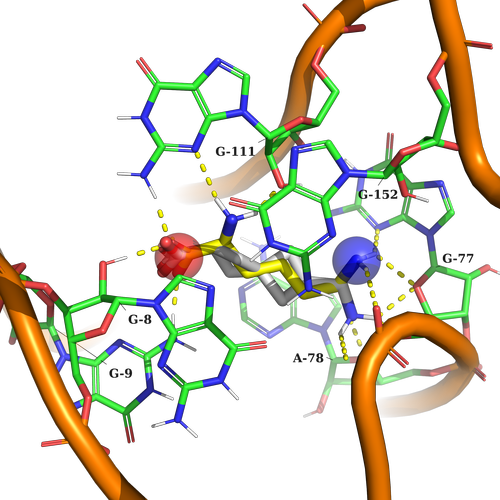
Example 2. In this case, the ligand should contain a string acceptor to imitate the COOH lysine group, and on the other part of the pocket NH3 group to crate caton-p interaction with surrounding nucleotides.