Guiding optimisation of fragment-derived lead compounds
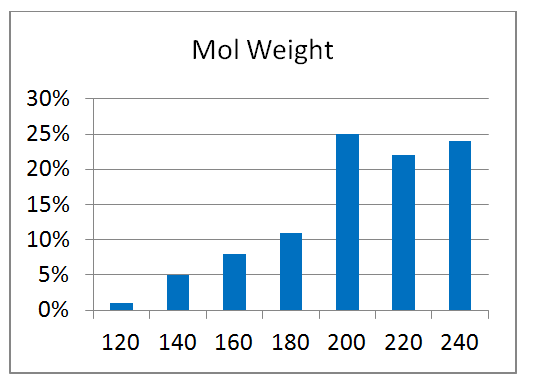
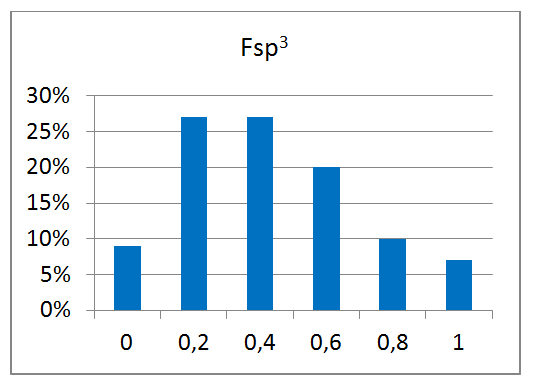
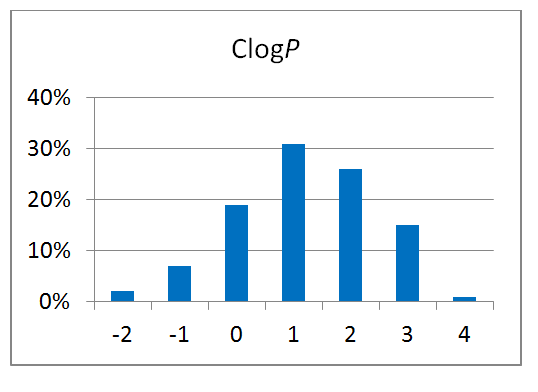
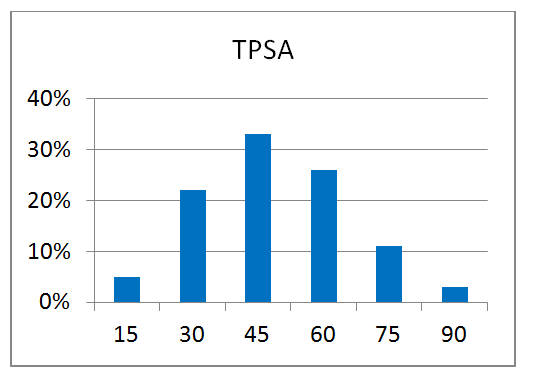
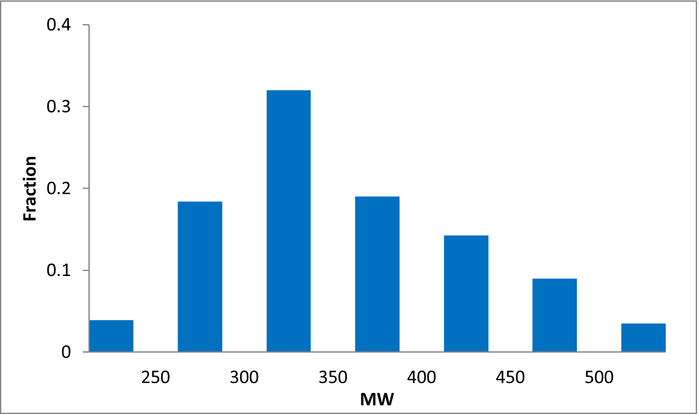
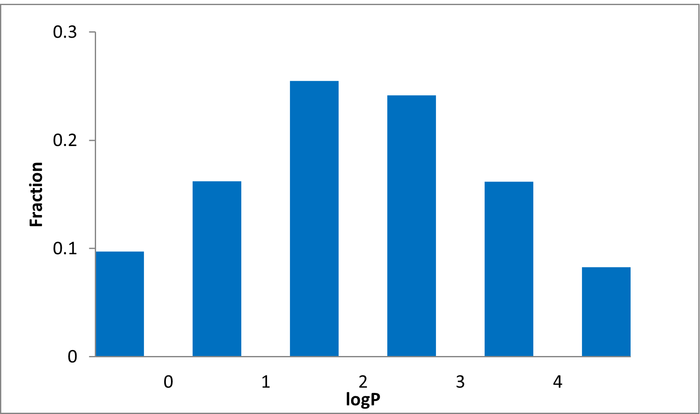
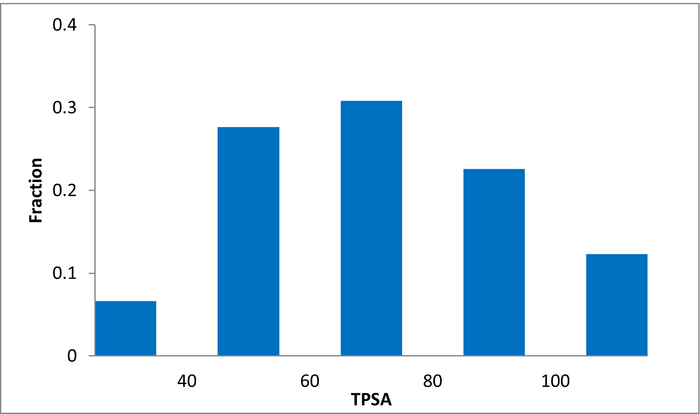
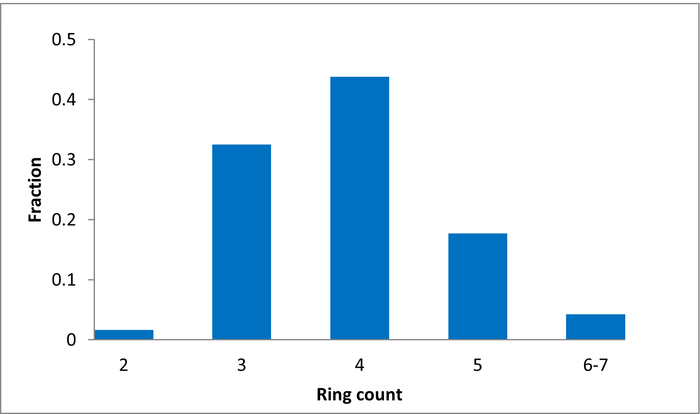
80 compounds
Reliable structural data of a target protein is the key to a successful drug discovery program. This knowledge is extremely important for the efficient development of selective and potent small molecule drugs. Recent achievements in the field of protein structure investigation are impressive and have enabled development of new approaches and screening techniques. Combination of modern, elaborated crystallographic methods with smart-design of chemical libraries can make a breakthrough in our understanding of protein structure changes and behavior.
Novel crystallographic screening methodology reported by O’Reilly et al. in Drug Discovery Today 2019 was developed at Astex Pharmaceuticals, Cambridge, UK. The high-concentration aqueous soaks were made with a chemically diverse and ultra-low-molecular-weight fragment library “MiniFrags” (heavy atom count 5–7). This allowed identification of hot and warm spots on proteins. High screening hit rates reflect enhanced sampling of chemical space. MiniFrag screening can represent thus a highly effective method for guiding optimisation of fragment-derived lead compounds.
We have collaborated with the Astex’ scientists on making MiniFrag library available to the research community in the most convenient format. Screening at 1M suggests that the fragments would be provided dry, ready for dissolution prior to protein soaking.
Typical Formats
Catalog No.
MiniFrag-80
Compounds
80
Amount
10 mg
Plates and formats
Dry samples formatted in individual sealed glass vials suitable for dissolution
Price
Catalog No.
MiniFrag-80
Compounds
80
Amount
Custom
Plates and formats
Any custom format
Price
Download SD file
Library design
The compounds in the MiniFrag Library offered by Enamine are identical to those proposed by Astex. Their structures and selection principles are described in O’Reilly et al. in Drug Discovery Today, Volume 24, Issue 5, May 2019, Pages 1081-1086. DOI: 10.1016/j.drudis.2019.03.009.
Designed for easy and rapid follow-up synthesis
860 compounds
Fragment screening is an efficient way of establishing initial points for drug discovery. However, seemingly simple small molecules don’t necessarily mean that their chemistry is not complex. Quite the contrary, fragment hits may be challenging to follow up. Synthesis routes can be long and rigid not allowing making small specific changes while growing, linking or merging the hit.
It was shown by Cox et al., in Chem. Sci. 2016 that the compounds synthesised from a robust and general synthetic reaction can be prioritized for building fragment library. Identification of so-called “poised” bonds in a fragment means that a library of analogues can be rapidly elaborated using standard parallel chemistry. First poised library was developed at Diamond Light Source, UK (Diamond) and Structural Genomic Consortium, UK (SGC) in the frame of one of the iNEXT Joint Research Activities. Enamine collaborated with the joint research alliance in design of a next generation of DSI-poised Library. DSI stands for Diamond, SGC and iNEXT. On January 10 2018 Diamond and SGC announced that Enamine would become a key supplier of poised fragment and analogue libraries to the XChem Facility in Oxford, UK.
Typical Formats
Catalog No.
DSI-860-10-Y-100
Compounds
8603 plates
Amount
10 µL of 100 mM DMSO solutions
Plates and formats
384-well plates Greiner Cat. No 781280, first two and last two columns empty, 320 compounds per plate
Price
Catalog No.
DSI-860-25-Y-100
Compounds
8603 plates
Amount
25 µL of 100 mM solutions in DMSO
Plates and formats
384-well plates, 1,2 & 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
DSI-860-3-Z-500
Compounds
8601 plate
Amount
3 µL of 500 mM solutions in DMSO-D6
Plates and formats
1536-well microplates, Cat. No LP-0400, 1-4 and 44-48 columns empty, 1280 compounds per plate
Price
Catalog No.
DSI-860-100-X-10
Compounds
86011 plates
Amount
100 µL of 10 mM DMSO solutions
Plates and formats
96-well plates, Greiner Cat. No 650201, round (U) bottom, 1 & 12 columns empty, 80 compounds per plate
Price
Catalog No.
DSI-860
Compounds
860
Amount
Custom
Plates and formats
Any custom format
Price
Download SD file
Library design
The original library developed by Diamond and SGC has been enhanced with a careful selection of fragments synthesized at Enamine via parallel chemistry from its building blocks, which are widely commercially available. Every researcher will be available to quickly synthesize analogues of the poised fragments because such building blocks are not complex and are exemplified with many analogues on the market. The reactions are frequently used in parallel chemistry: amide coupling, reductive amination, Suzuki-Miyaura coupling, aromatic nucleophilic substitution, synthesis of sulfonamides, ureas, etc.
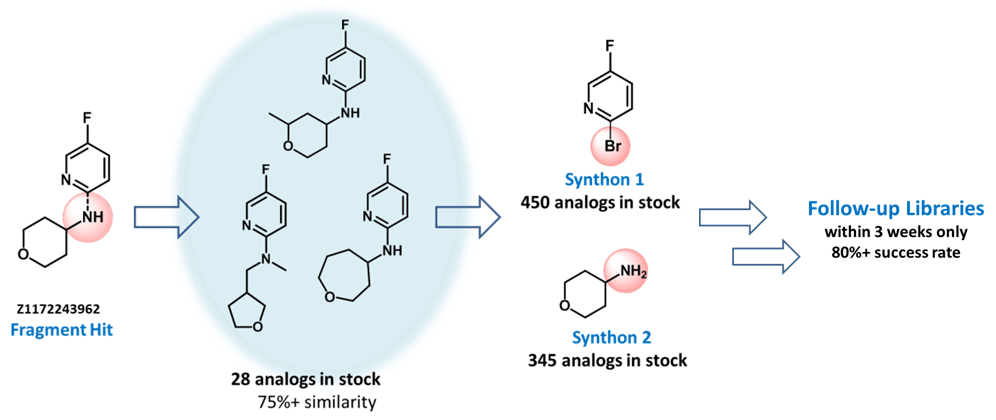
Examples of the molecules
Designed for discovery of new water channels modifiers
1 348 compounds
The library was designed to find molecules which target both end-chains of water channels (Aquaporins) AQ1, AQ4, AQ5 monomers and internal central pore of AQ5, that possess same physico-chemical properties as those in AQ1 and AQ4. A small number of known active inhibitors against this type of targets do not allowed us to build qualitative QSAR model or pharmacophore models. The HTS docking approach was used to search of new potential inhibitors, 2D similarity search was carried out to enrich the library with compounds similar to reported actives.
Download SD files
1 348 compounds for cherry-picking
Library Design
Docking optimization & calculations
Three protein structures (3D9S, 1IH5, 3GD8) were chosen after analysis of 11 available X-Ray structures (1H6I, 1IH5, 2ZZ9, 3GD8, 4NEF, 3D9S, 5DYE, etc.). However, it is known that shift in open/closed state is dramatically sensitive to some mutations. This data described several hot-spots, which are located inside the channel and were used for preparation of docking models on the primary stage of protein structure refinement, of course if these residues are involved in interaction, not in rigidity of the helix. Additionally the whole surface of each monomer was probed with FT-MAP server, which showed itself as a very accurate and trustworthy tool for binding site identification. It is based on evaluation of energy interaction between small organic probes (solvents, saturated and unsaturated cycles...) and the surface of interest.
The design of aquaporin library was performed with implementation of two different approaches: flexible docking and high throughput screening. Flexible docking protocol assumed a partially-free rotatable motion of side-chains, which are located in the entrance of both cavities in AQ5/AQ4/AQ1 proteins. The area of flexibility covered about 5 Å2 around the centre of the channel in one unit of the tetrameric structure. The centre of a Grid box (determination of the binding area) was selected based on residue mutation studies and FT-MAP calculations, mentioned above. To expand the volume of putative binding sites a set of real inhibitors, which were disclosed in publications and chemblDB, were docked together with compounds, which were shown to interact with the target (AqB013, Bumetanide, Zonisamide, Arbidol and Acetazolamide were taken from the literature and some neurotransmitters, like dopamine, epinephrine and serotonine, were chosen based on our collaboration with academicians.
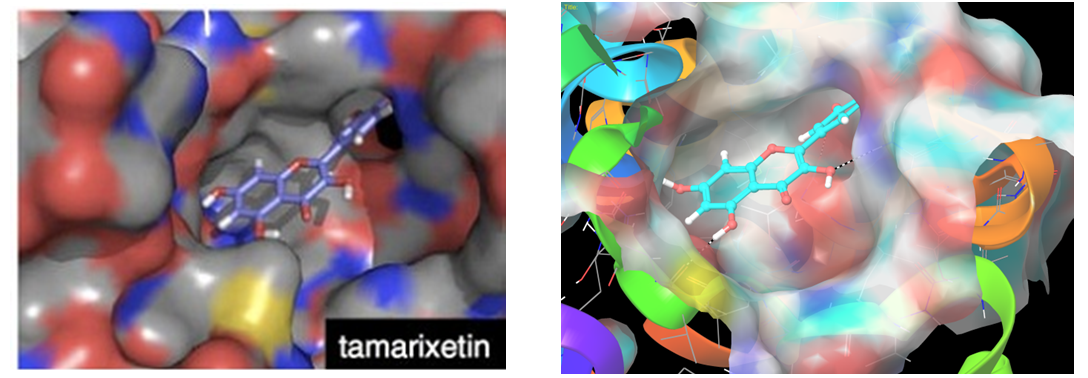
Reported binding mode of tamarixetin (left) and representation of hit molecule obtained after our docking calculations (right).
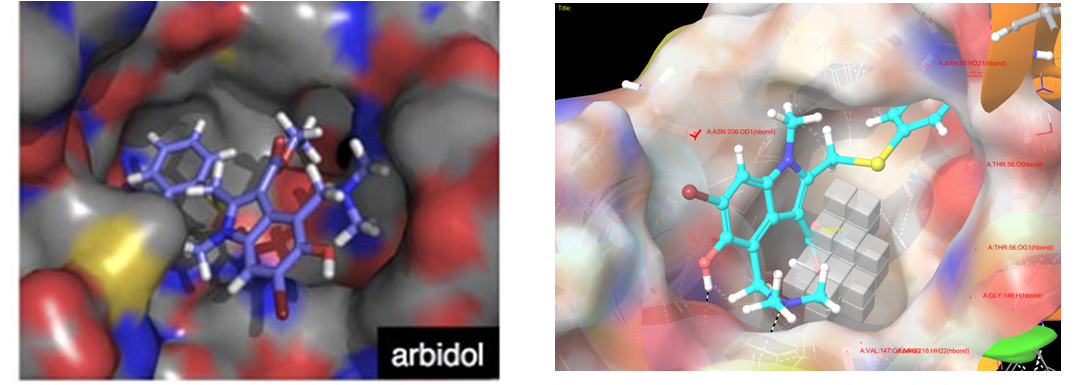
Several residues, which are responsible for substrate/inhibitor binding - G146, V147, R216, T56, (Asn58, Gly209, Val141), are also important for arbidol binding.
In this way a more suitable conformation of the gate was achieved in binding sites and these optimized structures were applied for screening procedure with slightly less exhaustive parameters of pose estimation and number of generated conformers. Here it is a short example of parametrization if the docking into the AQ5 model. Among constraints, which formed the filtering sieve, a hydrophobic core inside the channel and partial match against the set of amino acids (ARG:188, ASN:120, GLY:181, A:HIS:173, VAL:119 for AQ5 site2; and GLN:81, HIE:67, PRO:62, SER:152, THR:150, THR:150, TYR:90, TYR:90 for AQ5 site1), should be mentioned. All of them are shown in the figure. Those compounds which possessed a hydrophobic moiety and satisfied another condition (2-3 h-bond interactions with any of the listed residues), reached the next stage of more accurate and time-consuming docking procedure. Rating of these compounds is a combination of empiric values, post-processing calculation of energy, steric and h-bonding impacts. The more higher rating, the more probable binding of the ligand.
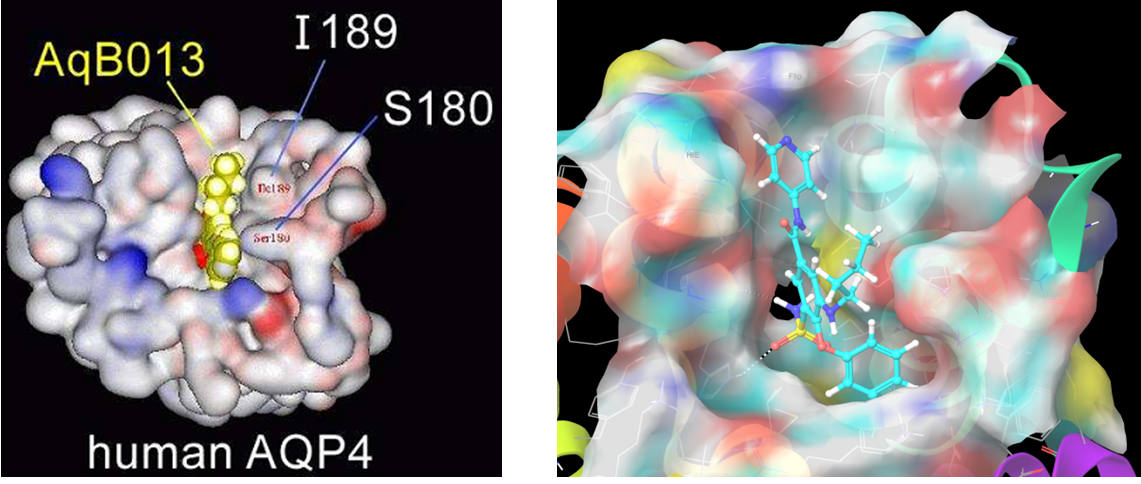
Compound AQB013 (the same for Bumetanide) docked in the second site of AQ4. The more preferable residues set is I189, S180, Phe175, Arg261, which are forming the opposite gate of the AQ4 channel.
Designed for discovery of novel BACE inhibitors
7 171 compounds
Alzheimer disease (AD) is one of the most difficult to treat CNS disorder. Despite of intensive investigation over last decades no effective medication has been developed against this horrible disease. However, the recent data on beta-secretase inhibition showed budding results in this field. BACE inhibition would seem to be a powerful tool against AD evolution in the early stages of the diseases. These data served as a basis for design of high-quality BACE targeted library with most diverse chemotypes and good CNS profile.
Download SD files
7 171 compounds for cherry-picking
Library Design
The database of activities related to BACE was carefully combined (as a result of searching in ChEMBL and literature data). In total 7 465 compounds with reported sufficient activity were found. All available PDB structures were analyzed as well. 23 most representative PDB complexes were used for molecular docking calculations. Further algorithm to search potential regulators of BACE target is presented below:
By using PDB, PDBe, pfam all available structures were selected. Also based on literature data and ChEMBL all active compounds were selected.
23 most representative structures were selected for molecular docking (2b8v, 2ohr, 2viz, 2wjo, 2zdz, 3exo, 3fkt, 3hw1, 3in3, 3l38, 3l5b, 3l5f, 3rsx, 3udy, 3uqw, 3vf3, 4acu, 4i10, 4l7h, 4xkx, 5hdv, 5kqf, 5uyu).
Then using obtained structures for BACE receptors and Enamine chemical DataBase flexible molecular docking.
As result for BACE protein target 7 171 (docking library) compounds were selected.
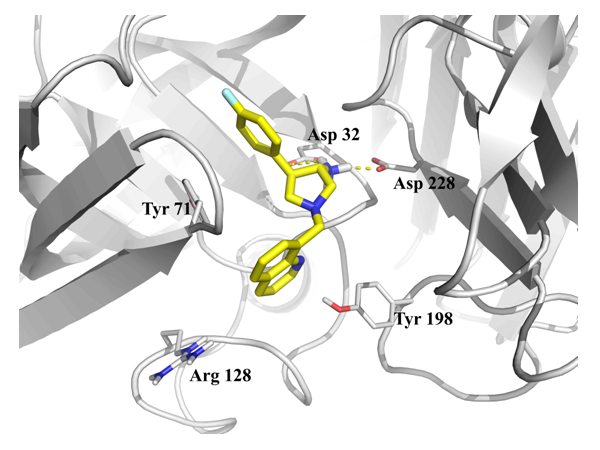
Fig. 1.Example of result of docking (4l7h).
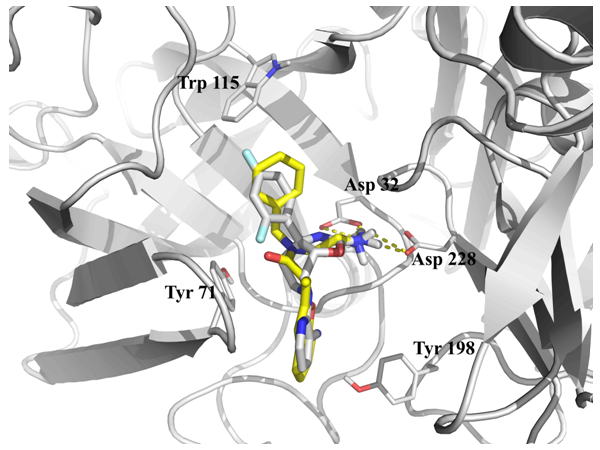
Fig. 2. Binding of 2ohr with native ligand (gray) and result of docking (Yellow).
Designed for identification of new actives against proteins essential for DNA stability
5 760 compounds
For a long time, targeting DNA was considered only in the context of non-specific and cytotoxic agents. Almost all known to date DNA-interacting drugs pose catastrophic actions to the live cells. Several well-known, previously, and still widely used cancer drugs such as Cisplatin, Doxorubicin, and ethidium bromide are non-selective DNA binders or intercalating compounds. Within the last several years protein-DNA interaction interfaces gained much attention among drug hunters. This area is under investigation and requires intensive research to bring new molecules to the clinic. To meet the growing need for new specific DNA binders, we have created a versatile library combining different approaches and tools.
The library has been plated for most convenient and quick access. Using our DNA focused library you receive multiple benefits, allowing you to save on time and costs in hit expansion and optimization:
- Analogs and hit samples resupply from dry stock of over 4.4M compounds
- Straightforward & low-cost synthesis of follow-up libraries through our REAL Database technology
- Medicinal chemistry support enhanced with on-site broad ADME/T panel
Typical Formats
DNA Library is available for supply in various pre-plated formats, including the following most popular ones:
Catalog No.
DNA-5760-0-Z-10
Compounds
5 760
5 plates
Amount
≤ 300 nL of 10 mM of DMSO solutions
Plates and formats
1536-well Echo LDV microplates, first and last four columns empty, 1280 compounds per plate
Price
Catalog No.
DNA-5760-10-Y-10
Compounds
5 760
18 plates
Amount
≤ 10 µL of 10 mM DMSO solutions
Plates and formats
384-well, Echo Qualified LDV microplates #001-12782 (LP-0200), first and last two columns empty, 320 compounds per plate
Price
Catalog No.
DNA-5760-50-Y-10
Compounds
5 760
18 plates
Amount
50 μL of 10 mM DMSO solutions
Plates and formats
384-well, Greiner Bio-One plates #781280, 1,2 and 23,24 columns empty, 320 compounds per plate
Price
Catalog No.
Library & follow-up package
Plates and formats
DNA-5760-10-Y-10 screening library 5 760 cmpds, hit resupply, analogs from 4.4M+ stock and synthesis from REAL Space
Price
*We will be happy to provide our library in any other most convenient for your project format. Please select among the following our standard microplates: Greiner Bio-One 781270, 784201, 781280, 651201 or Echo Qualified 001-12782 (LP-0200), 001-14555 (PP-0200), 001-6969 (LP-0400), C52621 or send your preferred labware. Compounds pooling can be provided upon request.
Download SD file
Library design
Different DNA structures have been carefully investigated and analyzed: (1) G-quadruplex structures; G-quadruplexes are noncanonical DNA structures that consist of stacked G-tetrads formed by guanine-rich sequences. These structures are stabilized by monovalent cations such as K+ and Na+. G-quadruplexes are commonly found in the promoter regions of oncogenes like MYC and play an important role in regulating gene expression. Understanding the binding of ligands to G-quadruplexes provides insights into potential therapeutic strategies for targeting oncogenes and modulating gene expression. (2) The double helix DNA structure is a fundamental arrangement where two strands of DNA wind around each other. The integrity of DNA double-strands is crucial for the stability and functioning of cells and organisms. DNA double-strand breaks are particularly critical events that can lead to various consequences. They are associated with the development of human syndromes, neurodegenerative diseases, immunodeficiency, and cancer.
To search for potential DNA ligands all available in PDB and in PDBe liganded DNA structures were analyzed. Then the following entries were selected for in silico screening: 5W77, 6JJ0, 7KBW, 5T4W, 6IJW, 7KWK, 5Z80, 6CCW, 6SX3, and 7EL7. The two main approaches, binding types, were used for search of potential ligands:
- Intercalation: ligands insert itself between the stacked DNA bases pairs.
- Binding insight the DNA grove: ligands binds to the DNA groove without significantly disrupting of the DNA structure.
The molecular docking simulations were conducted using the following queries: pharmacophore of H-bond acceptors and H-bond donors, aromatic group, stearic and volume exclusion features. Each pharmacophore model includes both common and specific features, making the binding points specific to each DNA target structure and its corresponding native ligand. Several examples are provided below.
Examples of molecular docking simulation to histone lysine methyltransferases
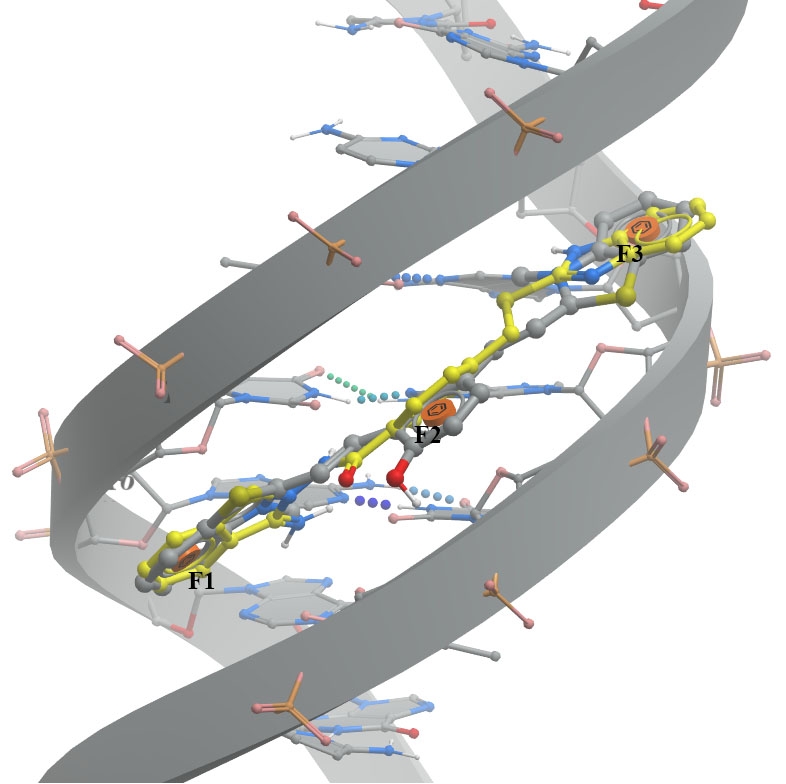
A. The minor groove (PDB ID 6IJW - left image) involves binding between QCy-DT DNA ligand and 6IJW, ligand should match with three aromatic features.
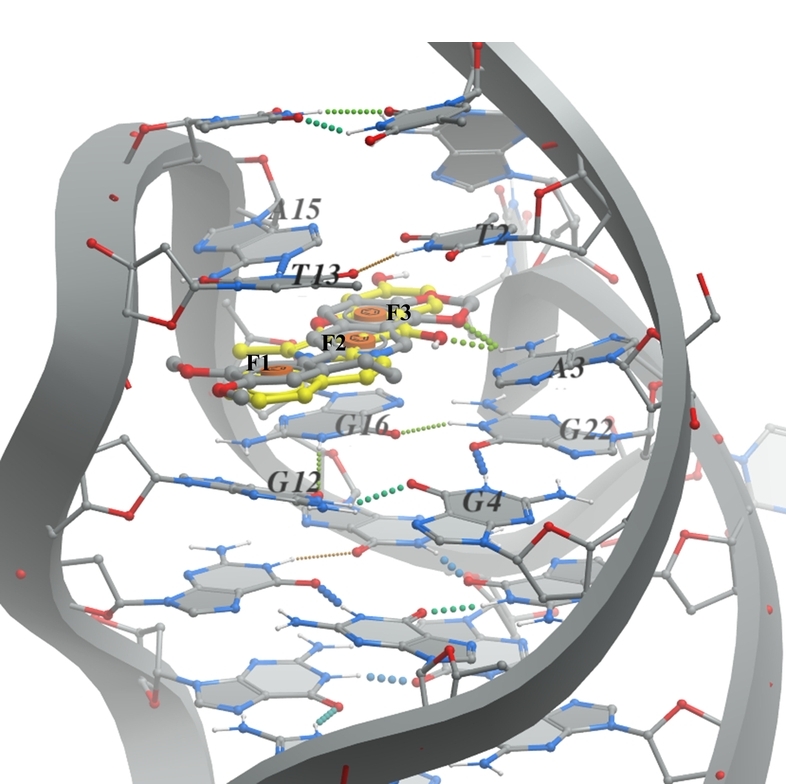
B. G-quadruplex (PDB ID 6CCW - right image) the pharmacophore model based on the presence of epiberberine intercalated within the G-quadruplex. The ligand should consist of three aromatic parts (F1, F2, and F3) that can stack between the DNA nucleobases G12, G16, A15, and T13.
DNA and its native ligand are colored in grey, docked ligands are shown in yellow.